Development of multivariable risk signature based on four immune-related RNA-binding proteins to predict survival and immune status in lung adenocarcinoma
- PMID: 36093548
- PMCID: PMC9459526
- DOI: 10.21037/tcr-22-698
Development of multivariable risk signature based on four immune-related RNA-binding proteins to predict survival and immune status in lung adenocarcinoma
Abstract
Background: This study aimed to construct a risk signature with predictive power based on immune-related RNA-binding proteins (RBPs) for lung adenocarcinoma (LUAD) patients.
Methods: The Cancer Genome Atlas (TCGA) database was used as the data source. Immune genes (IGs) were obtained from the Immunology Database and Analysis Portal (immPort) database. Differentially expressed RBPs and IGs between tumor and normal tissues were screened. For external validation, an independent cohort from the Gene-Expression Omnibus (GEO) database was used. The accuracy of the risk signature prediction was evaluated using Cox regression analysis and the receiver operating characteristic (ROC) curve.
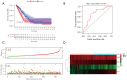
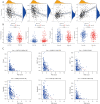
Results: The risk signature was constructed from four immune-related and prognostic RBPs (OAS3, PCF11, TLR7, and EXO1). The patients were divided into the low- and high-risk groups, with the low-risk group having a higher survival rate than the high-risk group. The risk signature outperformed other clinical parameters, with a multivariable hazard ratio of 1.862 (95% confidence interval: 1.292-2.683). The tumor immune microenvironment, stemness index, immune checkpoint, immune infiltration, and proportion of immune cells were significantly different between the low- and high-risk groups (all P<0.05).
Conclusions: The risk signature of immune-related RBPs can provide the basis for clinical decisions regarding diagnosis, prognosis, and immunotherapy in LUAD patients.
Keywords: Lung adenocarcinoma (LUAD); RNA-binding protein (RBP); drug sensitivity; immune infiltration; prognostic model.
2022 Translational Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-698/coif). The authors have no conflicts of interest to declare.
Figures





References
LinkOut - more resources
Full Text Sources
