C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni
- PMID: 36093987
- PMCID: PMC9631236
- DOI: 10.1021/acs.biochem.2c00364
C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni
Abstract
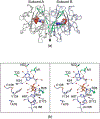
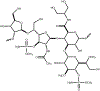
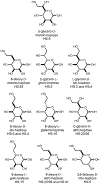
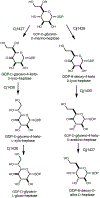
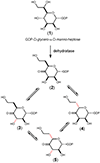
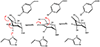
Campylobacter jejuni is a human pathogen and one of the leading causes of food poisoning in Europe and the United States. The outside of the bacterium is coated with a capsular polysaccharide that assists in the evasion of the host immune system. Many of the serotyped strains of C. jejuni contain a 6-deoxy-heptose moiety that is biosynthesized from GDP-d-glycero-d-manno-heptose by the successive actions of a 4,6-dehydratase, a C3/C5-epimerase, and a C4-reductase. We identified 18 different C3/C5-epimerases that could be clustered together into three groups at a sequence identity of >89%. Four of the enzymes from the largest cluster (from serotypes HS:3, HS:10, HS:23/36, and HS:41) were shown to only catalyze the epimerization at C3. Three enzymes from the second largest cluster (HS:2, HS:15, and HS:42) were shown to catalyze the epimerization at C3 and C5. Enzymes from the third cluster were not characterized. The three-dimensional structures of the epimerases from serotypes HS:3, HS:23/36, HS:15, and HS:41 were determined to resolutions of 1.5-1.9 Å. The overall subunit architecture places these enzymes into the diverse "cupin" superfamily. Within X-ray coordinate error, the immediate regions surrounding the active sites are identical, suggesting that factors extending farther out may influence product outcome. The X-ray crystal structures are consistent with His-67 and Tyr-134 acting as general acid/base catalysts for the epimerization of C3 and/or C5. Two amino acid changes (A76V/C136L) were enough to convert the C3-epimerase from serotype HS:3 to one that could now catalyze the epimerization at both C3 and C5.
Conflict of interest statement
The authors declare no competing conflicts of interests.
Figures



References
-
- Heimesaat MM, Backert S, Alter T, and Bereswill S (2021) Human Campylobacteriosis-A serious infectious threat in a One Health Perspective. Curr. Top. Microbiol. Immunol 431, 1–23. - PubMed
-
- Burnham PM, and Hendrixson DR (2018) Campylobacter jejuni: collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol 16, 551–565. - PubMed
-
- Kosma P (2008) Occurrence, synthesis and biosynthesis of bacterial heptoses. Curr. Org. Chem 12, 1021–1039.
-
- Karlyshev AV, Champion OL, Churcher C, Brisson JR, Jarrell HC, Gilbert M, Brochu D, St Michael F, Li JJ, Wakarchuk WW, Goodhead I, Sanders M, Stevens K, White B, Parkhill J, Wren BW, and Szymanski CM (2005) Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol 55, 90–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

