Limited Transmission of Klebsiella pneumoniae among Humans, Animals, and the Environment in a Caribbean Island, Guadeloupe (French West Indies)
- PMID: 36094181
- PMCID: PMC9603589
- DOI: 10.1128/spectrum.01242-22
Limited Transmission of Klebsiella pneumoniae among Humans, Animals, and the Environment in a Caribbean Island, Guadeloupe (French West Indies)
Abstract
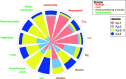
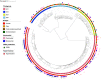
Guadeloupe (French West Indies), a Caribbean island, is an ideal place to study the reservoirs of the Klebsiella pneumoniae species complex (KpSC) and identify the routes of transmission between human and nonhuman sources due to its insularity, small population size, and small area. Here, we report an analysis of 590 biological samples, 546 KpSC isolates, and 331 genome sequences collected between January 2018 and May 2019. The KpSC appears to be common whatever the source. Extended-spectrum-β-lactamase (ESBL)-producing isolates (21.4%) belonged to K. pneumoniae sensu stricto (phylogroup Kp1), and all but one were recovered from the hospital setting. The distribution of species and phylogroups across the different niches was clearly nonrandom, with a distinct separation of Kp1 and Klebsiella variicola (Kp3). The most frequent sequence types (STs) (≥5 isolates) were previously recognized as high-risk multidrug-resistant (MDR) clones, namely, ST17, ST307, ST11, ST147, ST152, and ST45. Only 8 out of the 63 STs (12.7%) associated with human isolates were also found in nonhuman sources. A total of 22 KpSC isolates were defined as hypervirulent: 15 associated with human infections (9.8% of all human isolates), 4 (8.9%) associated with dogs, and 3 (15%) associated with pigs. Most of the human isolates (33.3%) belonged to the globally successful sublineage CG23-I. ST86 was the only clone shared by a human and a nonhuman (dog) source. Our work shows the limited transmission of KpSC isolates between human and nonhuman sources and points to the hospital setting as a cornerstone of the spread of MDR clones and antibiotic resistance genes. IMPORTANCE In this study, we characterized the presence and genomic features of isolates of the Klebsiella pneumoniae species complex (KpSC) from human and nonhuman sources in Guadeloupe (French West Indies) in order to identify the reservoirs and routes of transmission. This is the first study in an island environment, an ideal setting that limits the contribution of external imports. Our data showed the limited transmission of KpSC isolates between the different compartments. In contrast, we identified the hospital setting as the epicenter of antibiotic resistance due to the nosocomial spread of successful multidrug-resistant (MDR) K. pneumoniae clones and antibiotic resistance genes. Ecological barriers and/or limited exposure may restrict spread from the hospital setting to other reservoirs and vice versa. These results highlight the need for control strategies focused on health care centers, using genomic surveillance to limit the spread, particularly of high-risk clones, of this important group of MDR pathogens.
Keywords: Caribbean; ESBL; Klebsiella pneumoniae; One Health; genomic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Multidrug-resistant ESBL-producing Klebsiella pneumoniae complex in Czech hospitals, wastewaters and surface waters.Antimicrob Resist Infect Control. 2024 Nov 26;13(1):141. doi: 10.1186/s13756-024-01496-0. Antimicrob Resist Infect Control. 2024. PMID: 39593189 Free PMC article.
-
High Prevalence of Klebsiella pneumoniae in European Food Products: a Multicentric Study Comparing Culture and Molecular Detection Methods.Microbiol Spectr. 2022 Feb 23;10(1):e0237621. doi: 10.1128/spectrum.02376-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196810 Free PMC article.
-
Comprehensive study reveals phenotypic heterogeneity in Klebsiella pneumoniae species complex isolates.Sci Rep. 2024 Mar 11;14(1):5876. doi: 10.1038/s41598-024-55546-z. Sci Rep. 2024. PMID: 38467675 Free PMC article.
-
Identification and characterization of CTX-M-15 producing Klebsiella pneumoniae clone ST101 in a Hungarian university teaching hospital.Acta Microbiol Immunol Hung. 2015 Sep;62(3):233-45. doi: 10.1556/030.62.2015.3.2. Acta Microbiol Immunol Hung. 2015. PMID: 26551567 Review.
-
From soil to surface water: exploring Klebsiella 's clonal lineages and antibiotic resistance odyssey in environmental health.BMC Microbiol. 2025 Feb 27;25(1):97. doi: 10.1186/s12866-025-03798-8. BMC Microbiol. 2025. PMID: 40012032 Free PMC article. Review.
Cited by
-
Do Microorganisms in Bathing Water in Guadeloupe (French West Indies) Have Resistance Genes?Antibiotics (Basel). 2024 Jan 16;13(1):87. doi: 10.3390/antibiotics13010087. Antibiotics (Basel). 2024. PMID: 38247646 Free PMC article.
-
Genetic characterization and clonal analysis of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae from canine and human origins.Front Vet Sci. 2024 Nov 25;11:1464934. doi: 10.3389/fvets.2024.1464934. eCollection 2024. Front Vet Sci. 2024. PMID: 39654836 Free PMC article.
-
Occurrence and characteristics of extended-spectrum-β-lactamase- and pAmpC-producing Klebsiella pneumoniae isolated from companion animals with urinary tract infections.PLoS One. 2024 Jan 16;19(1):e0296709. doi: 10.1371/journal.pone.0296709. eCollection 2024. PLoS One. 2024. PMID: 38227590 Free PMC article.
-
Triplex real-time PCR ZKIR-T assay for simultaneous detection of the Klebsiella pneumoniae species complex and identification of K. pneumoniae sensu stricto.Microbiol Spectr. 2024 Oct 22;12(12):e0033624. doi: 10.1128/spectrum.00336-24. Online ahead of print. Microbiol Spectr. 2024. PMID: 39436128 Free PMC article.
-
Environmental Exposures and the Human Gut Resistome in Northwest Ecuador.medRxiv [Preprint]. 2025 May 23:2025.05.23.25327954. doi: 10.1101/2025.05.23.25327954. medRxiv. 2025. PMID: 40475154 Free PMC article. Preprint.
References
-
- Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M, Ouakrim DA, Oliveira TC, Struelens MJ, Suetens C, Monnet DL, Burden of AMR Collaborative Group . 2019. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis 19:56–66. doi:10.1016/S1473-3099(18)30605-4. - DOI - PMC - PubMed
-
- Breurec S, Bouchiat C, Sire JM, Moquet O, Bercion R, Cisse MF, Glaser P, Ndiaye O, Ka S, Salord H, Seck A, Sy HS, Michel R, Garin B. 2016. High third-generation cephalosporin resistant Enterobacteriaceae prevalence rate among neonatal infections in Dakar, Senegal. BMC Infect Dis 16:587. doi:10.1186/s12879-016-1935-y. - DOI - PMC - PubMed
-
- Wyres KL, Wick RR, Judd LM, Froumine R, Tokolyi A, Gorrie CL, Lam MMC, Duchene S, Jenney A, Holt KE. 2019. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet 15:e1008114. doi:10.1371/journal.pgen.1008114. - DOI - PMC - PubMed
-
- Rodrigues C, Passet V, Rakotondrasoa A, Diallo TA, Criscuolo A, Brisse S. 2019. Description of Klebsiella africanensis sp. nov., Klebsiella variicola subsp. tropicalensis subsp. nov. and Klebsiella variicola subsp. variicola subsp. nov. Res Microbiol 170:165–170. doi:10.1016/j.resmic.2019.02.003. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

