HIV-1 Preintegration Complex Preferentially Integrates the Viral DNA into Nucleosomes Containing Trimethylated Histone 3-Lysine 36 Modification and Flanking Linker DNA
- PMID: 36094316
- PMCID: PMC9517705
- DOI: 10.1128/jvi.01011-22
HIV-1 Preintegration Complex Preferentially Integrates the Viral DNA into Nucleosomes Containing Trimethylated Histone 3-Lysine 36 Modification and Flanking Linker DNA
Abstract
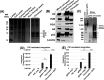
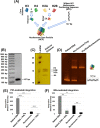
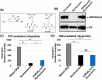
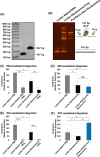
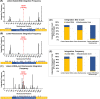
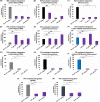
HIV-1 DNA is preferentially integrated into chromosomal hot spots by the preintegration complex (PIC). To understand the mechanism, we measured the DNA integration activity of PICs-extracted from infected cells-and intasomes, biochemically assembled PIC substructures using a number of relevant target substrates. We observed that PIC-mediated integration into human chromatin is preferred compared to genomic DNA. Surprisingly, nucleosomes lacking histone modifications were not preferred integration compared to the analogous naked DNA. Nucleosomes containing the trimethylated histone 3 lysine 36 (H3K36me3), an epigenetic mark linked to active transcription, significantly stimulated integration, but the levels remained lower than the naked DNA. Notably, H3K36me3-modified nucleosomes with linker DNA optimally supported integration mediated by the PIC but not by the intasome. Interestingly, optimal intasome-mediated integration required the cellular cofactor LEDGF. Unexpectedly, LEDGF minimally affected PIC-mediated integration into naked DNA but blocked integration into nucleosomes. The block for the PIC-mediated integration was significantly relieved by H3K36me3 modification. Mapping the integration sites in the preferred substrates revealed that specific features of the nucleosome-bound DNA are preferred for integration, whereas integration into naked DNA was random. Finally, biochemical and genetic studies demonstrate that DNA condensation by the H1 protein dramatically reduces integration, providing further evidence that features inherent to the open chromatin are preferred for HIV-1 integration. Collectively, these results identify the optimal target substrate for HIV-1 integration, report a mechanistic link between H3K36me3 and integration preference, and importantly, reveal distinct mechanisms utilized by the PIC for integration compared to the intasomes. IMPORTANCE HIV-1 infection is dependent on integration of the viral DNA into the host chromosomes. The preintegration complex (PIC) containing the viral DNA, the virally encoded integrase (IN) enzyme, and other viral/host factors carries out HIV-1 integration. HIV-1 integration is not dependent on the target DNA sequence, and yet the viral DNA is selectively inserted into specific "hot spots" of human chromosomes. A growing body of literature indicates that structural features of the human chromatin are important for integration targeting. However, the mechanisms that guide the PIC and enable insertion of the PIC-associated viral DNA into specific hot spots of the human chromosomes are not fully understood. In this study, we describe a biochemical mechanism for the preference of the HIV-1 DNA integration into open chromatin. Furthermore, our study defines a direct role for the histone epigenetic mark H3K36me3 in HIV-1 integration preference and identify an optimal substrate for HIV-1 PIC-mediated viral DNA integration.
Keywords: H3K36me3; HIV; PIC; PTM; chromatin; histone; human immunodeficiency virus; intasome; integration; nucleosome; posttranslational modification; preintegration complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


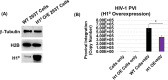
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI136740/AI/NIAID NIH HHS/United States
- R01 AI143649/AI/NIAID NIH HHS/United States
- G12 MD007586/MD/NIMHD NIH HHS/United States
- R01 AI162694/AI/NIAID NIH HHS/United States
- R24 DA036420/DA/NIDA NIH HHS/United States
- R56 AI122960/AI/NIAID NIH HHS/United States
- R01 DA057204/DA/NIDA NIH HHS/United States
- F31 AI150488/AI/NIAID NIH HHS/United States
- R01 DA042348/DA/NIDA NIH HHS/United States
- R25 GM059994/GM/NIGMS NIH HHS/United States
- R25 AI164610/AI/NIAID NIH HHS/United States
- R01 AI170228/AI/NIAID NIH HHS/United States
- U54 MD007586/MD/NIMHD NIH HHS/United States
- P30 AI110527/AI/NIAID NIH HHS/United States
- U54 MD007593/MD/NIMHD NIH HHS/United States
LinkOut - more resources
Full Text Sources

