Impaired IL-23-dependent induction of IFN-γ underlies mycobacterial disease in patients with inherited TYK2 deficiency
- PMID: 36094518
- PMCID: PMC9472563
- DOI: 10.1084/jem.20220094
Impaired IL-23-dependent induction of IFN-γ underlies mycobacterial disease in patients with inherited TYK2 deficiency
Abstract
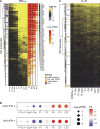
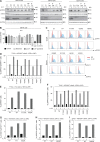
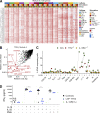
Human cells homozygous for rare loss-of-expression (LOE) TYK2 alleles have impaired, but not abolished, cellular responses to IFN-α/β (underlying viral diseases in the patients) and to IL-12 and IL-23 (underlying mycobacterial diseases). Cells homozygous for the common P1104A TYK2 allele have selectively impaired responses to IL-23 (underlying isolated mycobacterial disease). We report three new forms of TYK2 deficiency in six patients from five families homozygous for rare TYK2 alleles (R864C, G996R, G634E, or G1010D) or compound heterozygous for P1104A and a rare allele (A928V). All these missense alleles encode detectable proteins. The R864C and G1010D alleles are hypomorphic and loss-of-function (LOF), respectively, across signaling pathways. By contrast, hypomorphic G996R, G634E, and A928V mutations selectively impair responses to IL-23, like P1104A. Impairment of the IL-23-dependent induction of IFN-γ is the only mechanism of mycobacterial disease common to patients with complete TYK2 deficiency with or without TYK2 expression, partial TYK2 deficiency across signaling pathways, or rare or common partial TYK2 deficiency specific for IL-23 signaling.
© 2022 Ogishi et al.
Conflict of interest statement
Disclosures: The authors declare no competing financial interests.
Figures










References
-
- Arias, A.A., Perez-Velez C.M., Orrego J.C., Moncada-Velez M., Rojas J.L., Wilches A., Restrepo A., Trujillo M., Garces C., Arango-Ferreira C., et al. 2017. Severe enteropathy and hypogammaglobulinemia complicating refractory mycobacterium tuberculosis complex disseminated disease in a child with IL-12Rbeta1 deficiency. J. Clin. Immunol. 37:732–738. 10.1007/s10875-017-0435-1 - DOI - PubMed
-
- Belkadi, A., Pedergnana V., Cobat A., Itan Y., Vincent Q.B., Abhyankar A., Shang L., El Baghdadi J., Bousfiha A., Exome/Array Consortium, et al. 2016. Whole-exome sequencing to analyze population structure, parental inbreeding, and familial linkage. Proc. Natl. Acad. Sci. USA. 113:6713–6718. 10.1073/pnas.1606460113 - DOI - PMC - PubMed

