Calculation of Heat Capacity Changes in Enzyme Catalysis and Ligand Binding
- PMID: 36094903
- PMCID: PMC9558309
- DOI: 10.1021/acs.jctc.2c00646
Calculation of Heat Capacity Changes in Enzyme Catalysis and Ligand Binding
Abstract
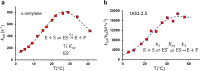
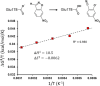
It has been suggested that heat capacity changes in enzyme catalysis may be the underlying reason for temperature optima that are not related to unfolding of the enzyme. If this were to be a common phenomenon, it would have major implications for our interpretation of enzyme kinetics. In most cases, the support for the possible existence of a nonzero (negative) activation heat capacity, however, only relies on fitting such a kinetic model to experimental data. It is therefore of fundamental interest to try to use computer simulations to address this issue. One way is simply to calculate the temperature dependence of the activation free energy and determine whether the relationship is linear or not. An alternative approach is to calculate the absolute heat capacities of the reactant and transition states from plain molecular dynamics simulations using either the temperature derivative or fluctuation formula for the enthalpy. Here, we examine these different approaches for a designer enzyme with a temperature optimum that is not caused by unfolding. Benchmark calculations for the heat capacity of liquid water are first carried out using different thermostats. It is shown that the derivative formula for the heat capacity is generally the most robust and insensitive to the thermostat used and its parameters. The enzyme calculations using this method give results in agreement with direct calculations of activation free energies and show no sign of a negative activation heat capacity. We also provide a simple scheme for the calculation of binding heat capacity changes, which is of clear interest in ligand design, and demonstrate it for substrate binding to the designer enzyme. Neither in that case do the simulations predict any negative heat capacity change.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
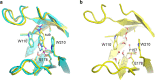

Similar articles
-
Computational Analysis of Heat Capacity Effects in Protein-Ligand Binding.J Chem Theory Comput. 2024 Jul 9;20(13):5708-5716. doi: 10.1021/acs.jctc.4c00525. Epub 2024 Jun 13. J Chem Theory Comput. 2024. PMID: 38870420 Free PMC article.
-
Hidden Conformational States and Strange Temperature Optima in Enzyme Catalysis.Biochemistry. 2020 Oct 13;59(40):3844-3855. doi: 10.1021/acs.biochem.0c00705. Epub 2020 Sep 25. Biochemistry. 2020. PMID: 32975950 Free PMC article.
-
Thermodynamics of protein-ligand interactions: history, presence, and future aspects.J Recept Signal Transduct Res. 2004 Feb;24(1-2):1-52. doi: 10.1081/rrs-120037896. J Recept Signal Transduct Res. 2004. PMID: 15344878 Review.
-
Heat Capacity Changes for Transition-State Analogue Binding and Catalysis with Human 5'-Methylthioadenosine Phosphorylase.ACS Chem Biol. 2017 Feb 17;12(2):464-473. doi: 10.1021/acschembio.6b00885. Epub 2016 Dec 27. ACS Chem Biol. 2017. PMID: 28026167 Free PMC article.
-
On the link between conformational changes, ligand binding and heat capacity.Biochim Biophys Acta. 2016 May;1860(5):868-878. doi: 10.1016/j.bbagen.2015.10.010. Epub 2015 Oct 22. Biochim Biophys Acta. 2016. PMID: 26476135 Review.
Cited by
-
Interplay of structural preorganization and conformational sampling in UDP-glucuronic acid 4-epimerase catalysis.Nat Commun. 2024 May 8;15(1):3897. doi: 10.1038/s41467-024-48281-6. Nat Commun. 2024. PMID: 38719841 Free PMC article.
-
A structural perspective on the temperature-dependent activity of enzymes.bioRxiv [Preprint]. 2024 Aug 23:2024.08.23.609221. doi: 10.1101/2024.08.23.609221. bioRxiv. 2024. Update in: Structure. 2025 May 1;33(5):924-934.e2. doi: 10.1016/j.str.2025.02.013. PMID: 39229032 Free PMC article. Updated. Preprint.
-
Computer Simulations of the Temperature Dependence of Enzyme Reactions.J Chem Theory Comput. 2025 Feb 11;21(3):1017-1028. doi: 10.1021/acs.jctc.4c01733. Epub 2025 Jan 30. J Chem Theory Comput. 2025. PMID: 39884967 Free PMC article. Review.
-
Computational design of the temperature optimum of an enzyme reaction.Sci Adv. 2023 Jun 28;9(26):eadi0963. doi: 10.1126/sciadv.adi0963. Epub 2023 Jun 28. Sci Adv. 2023. PMID: 37379391 Free PMC article.
-
Cooperative Conformational Transitions Underpin the Activation Heat Capacity in the Temperature Dependence of Enzyme Catalysis.ACS Catal. 2024 Mar 8;14(7):4379-4394. doi: 10.1021/acscatal.3c05584. eCollection 2024 Apr 5. ACS Catal. 2024. PMID: 38633402 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
