SIRT5 is a proviral factor that interacts with SARS-CoV-2 Nsp14 protein
- PMID: 36095012
- PMCID: PMC9499238
- DOI: 10.1371/journal.ppat.1010811
SIRT5 is a proviral factor that interacts with SARS-CoV-2 Nsp14 protein
Abstract
SARS-CoV-2 non-structural protein Nsp14 is a highly conserved enzyme necessary for viral replication. Nsp14 forms a stable complex with non-structural protein Nsp10 and exhibits exoribonuclease and N7-methyltransferase activities. Protein-interactome studies identified human sirtuin 5 (SIRT5) as a putative binding partner of Nsp14. SIRT5 is an NAD-dependent protein deacylase critical for cellular metabolism that removes succinyl and malonyl groups from lysine residues. Here we investigated the nature of this interaction and the role of SIRT5 during SARS-CoV-2 infection. We showed that SIRT5 interacts with Nsp14, but not with Nsp10, suggesting that SIRT5 and Nsp10 are parts of separate complexes. We found that SIRT5 catalytic domain is necessary for the interaction with Nsp14, but that Nsp14 does not appear to be directly deacylated by SIRT5. Furthermore, knock-out of SIRT5 or treatment with specific SIRT5 inhibitors reduced SARS-CoV-2 viral levels in cell-culture experiments. SIRT5 knock-out cells expressed higher basal levels of innate immunity markers and mounted a stronger antiviral response, independently of the Mitochondrial Antiviral Signaling Protein MAVS. Our results indicate that SIRT5 is a proviral factor necessary for efficient viral replication, which opens novel avenues for therapeutic interventions.
Conflict of interest statement
The Krogan Laboratory has received research support from Vir Biotechnology, F. Hoffmann-La Roche, and Rezo Therapeutics. Nevan J. Krogan has financially compensated consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics, Interline Therapeutics, Rezo Therapeutics, GEn1E Lifesciences, Inc. and Twist Bioscience Corp. He is on the Board of Directors of Rezo Therapeutics and is a shareholder in Tenaya Therapeutics, Maze Therapeutics, Rezo Therapeutics, and Interline Therapeutics.
Figures
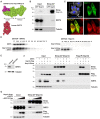





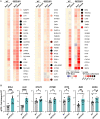

Update of
-
SIRT5 is a proviral factor that interacts with SARS-CoV-2 Nsp14 protein.bioRxiv [Preprint]. 2022 Jan 5:2022.01.04.474979. doi: 10.1101/2022.01.04.474979. bioRxiv. 2022. Update in: PLoS Pathog. 2022 Sep 12;18(9):e1010811. doi: 10.1371/journal.ppat.1010811. PMID: 35018374 Free PMC article. Updated. Preprint.
References
-
- Laurent EMN, Sofianatos Y, Komarova A, Gimeno JP. Global BioID-based SARS-CoV-2 proteins proximal interactome unveils novel ties between viral polypeptides and host factors involved in multiple COVID19 …. BioRxiv. 2020. Available: https://www.biorxiv.org/content/10.1101/2020.08.28.272955v1.abstract - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

