Novel viral splicing events and open reading frames revealed by long-read direct RNA sequencing of adenovirus transcripts
- PMID: 36095031
- PMCID: PMC9499273
- DOI: 10.1371/journal.ppat.1010797
Novel viral splicing events and open reading frames revealed by long-read direct RNA sequencing of adenovirus transcripts
Abstract
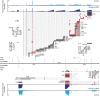
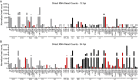
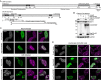
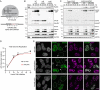
Adenovirus is a common human pathogen that relies on host cell processes for transcription and processing of viral RNA and protein production. Although adenoviral promoters, splice junctions, and polyadenylation sites have been characterized using low-throughput biochemical techniques or short read cDNA-based sequencing, these technologies do not fully capture the complexity of the adenoviral transcriptome. By combining Illumina short-read and nanopore long-read direct RNA sequencing approaches, we mapped transcription start sites and RNA cleavage and polyadenylation sites across the adenovirus genome. In addition to confirming the known canonical viral early and late RNA cassettes, our analysis of splice junctions within long RNA reads revealed an additional 35 novel viral transcripts that meet stringent criteria for expression. These RNAs include fourteen new splice junctions which lead to expression of canonical open reading frames (ORFs), six novel ORF-containing transcripts, and 15 transcripts encoding for messages that could alter protein functions through truncation or fusion of canonical ORFs. In addition, we detect RNAs that bypass canonical cleavage sites and generate potential chimeric proteins by linking distinct gene transcription units. Among these chimeric proteins we detected an evolutionarily conserved protein containing the N-terminus of E4orf6 fused to the downstream DBP/E2A ORF. Loss of this novel protein, E4orf6/DBP, was associated with aberrant viral replication center morphology and poor viral spread. Our work highlights how long-read sequencing technologies combined with mass spectrometry can reveal further complexity within viral transcriptomes and resulting proteomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Berk AJ. Adenoviridae. 6th ed. In: Knipe David M., Howley Peter M., editors. Fields Virology. 6th ed. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2013. pp. 1704–1731.
Publication types
MeSH terms
Substances
Grants and funding
- R21 AI147163/AI/NIAID NIH HHS/United States
- P01 CA196539/CA/NCI NIH HHS/United States
- R21 AI130618/AI/NIAID NIH HHS/United States
- K99 AI159049/AI/NIAID NIH HHS/United States
- R01 AI145266/AI/NIAID NIH HHS/United States
- R01 AI118891/AI/NIAID NIH HHS/United States
- R21 AI154654/AI/NIAID NIH HHS/United States
- R01 AI152543/AI/NIAID NIH HHS/United States
- F32 AI138432/AI/NIAID NIH HHS/United States
- R01 HD106051/HD/NICHD NIH HHS/United States
- T32 CA115299/CA/NCI NIH HHS/United States
- R01 AI121321/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources

