Apicoplast biogenesis mediated by ATG8 requires the ATG12-ATG5-ATG16L and SNAP29 complexes in Toxoplasma gondii
- PMID: 36095096
- PMCID: PMC10012919
- DOI: 10.1080/15548627.2022.2123639
Apicoplast biogenesis mediated by ATG8 requires the ATG12-ATG5-ATG16L and SNAP29 complexes in Toxoplasma gondii
Abstract
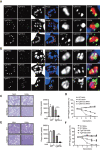
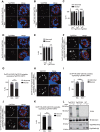
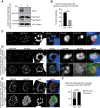
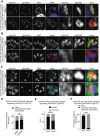
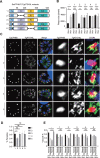
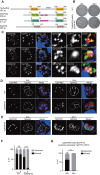
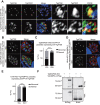
In apicomplexan parasites, the macroautophagy/autophagy machinery is repurposed to maintain the plastid-like organelle apicoplast. Previously, we showed that in Toxoplasma and Plasmodium, ATG12 interacts with ATG5 in a non-covalent manner, in contrast to the covalent interaction in most organisms. However, it remained unknown whether apicomplexan parasites have functional orthologs of ATG16L1, a protein that is essential for the function of the covalent ATG12-ATG5 complex in vivo in other organisms. Furthermore, the mechanism used by the autophagy machinery to maintain the apicoplast is unclear. We report that the ATG12-ATG5-ATG16L complex exists in Toxoplasma gondii (Tg). This complex is localized on isolated structures at the periphery of the apicoplast dependent on TgATG16L. Inducible depletion of TgATG12, TgATG5, or TgATG16L caused loss of the apicoplast and affected parasite growth. We found that a putative soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) protein, synaptosomal-associated protein 29 (TgSNAP29, Qbc SNARE), is required to maintain the apicoplast in T. gondii. TgSNAP29 depletion disrupted TgATG8 localization at the apicoplast. Additionally, we identified a putative ubiquitin-interacting motif-docking site (UDS) of TgATG8. Mutation of the UDS site abolished TgATG8 localization on the apicoplast but not lipidation. These findings suggest that the TgATG12-TgATG5-TgATG16L complex is required for biogenesis of the apicoplast, in which TgATG8 is translocated to the apicoplast via vesicles in a SNARE -dependent manner in T. gondii.Abbreviations: AID: auxin-inducible degron; CCD: coiled-coil domain; HFF: human foreskin fibroblast; IAA: indole-3-acetic acid; LAP: LC3-associated phagocytosis; NAA: 1-naphthaleneacetic acid; PtdIns3P: phosphatidylinositol-3-phosphate; SNARE: soluble N-ethylmaleimide sensitive factor attachment protein receptor; UDS: ubiquitin-interacting motif-docking site; UIM: ubiquitin-interacting motif.
Keywords: Apicoplast; T. gondii; TgATG12–TgATG5-TgATG16L; TgATG8; TgSNAP29; ubiquitin-interacting motif.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


Similar articles
-
Toxoplasma TgAtg8-TgAtg3 Interaction Primarily Contributes to Apicoplast Inheritance and Parasite Growth in Tachyzoite.Microbiol Spectr. 2022 Feb 23;10(1):e0149521. doi: 10.1128/spectrum.01495-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196797 Free PMC article.
-
Autophagy-Related Protein ATG8 Has a Noncanonical Function for Apicoplast Inheritance in Toxoplasma gondii.mBio. 2015 Oct 27;6(6):e01446-15. doi: 10.1128/mBio.01446-15. mBio. 2015. PMID: 26507233 Free PMC article.
-
Toxoplasma gondii autophagy-related protein ATG7 maintains apicoplast inheritance by stabilizing and lipidating ATG8.Biochim Biophys Acta Mol Basis Dis. 2024 Jan;1870(1):166891. doi: 10.1016/j.bbadis.2023.166891. Epub 2023 Sep 20. Biochim Biophys Acta Mol Basis Dis. 2024. PMID: 37739091
-
ATG8 localization in apicomplexan parasites: apicoplast and more?Autophagy. 2014 Sep;10(9):1487-94. doi: 10.4161/auto.32183. Epub 2014 Aug 4. Autophagy. 2014. PMID: 25102412 Free PMC article. Review.
-
The metabolic pathways and transporters of the plastid organelle in Apicomplexa.Curr Opin Microbiol. 2021 Oct;63:250-258. doi: 10.1016/j.mib.2021.07.016. Epub 2021 Aug 26. Curr Opin Microbiol. 2021. PMID: 34455306 Review.
Cited by
-
Discovery of Evolutionary Loss of the Ubiquitin-like Autophagy-Related ATG12 System in a Lineage of Apicomplexa.Cells. 2025 Jan 15;14(2):121. doi: 10.3390/cells14020121. Cells. 2025. PMID: 39851549 Free PMC article.
-
Autophagy genes in biology and disease.Nat Rev Genet. 2023 Jun;24(6):382-400. doi: 10.1038/s41576-022-00562-w. Epub 2023 Jan 12. Nat Rev Genet. 2023. PMID: 36635405 Free PMC article. Review.
-
A kalihinol analog disrupts apicoplast function and vesicular trafficking in P. falciparum malaria.Science. 2024 Sep 27;385(6716):eadm7966. doi: 10.1126/science.adm7966. Epub 2024 Sep 27. Science. 2024. PMID: 39325875
-
Chinese medicine monomers for hepatocellular carcinoma: New ideas related to autophagy.World J Gastroenterol. 2025 Jul 14;31(26):106113. doi: 10.3748/wjg.v31.i26.106113. World J Gastroenterol. 2025. PMID: 40678709 Free PMC article. Review.
-
TgATG9 is required for autophagosome biogenesis and maintenance of chronic infection in Toxoplasma gondii.Autophagy Rep. 2024;3(1):2418256. doi: 10.1080/27694127.2024.2418256. Epub 2024 Oct 23. Autophagy Rep. 2024. PMID: 39600488 Free PMC article.
References
-
- Mizushima N, Yoshimori T, Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27(1):107–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
