Sibling rivalry among the ZBTB transcription factor family: homodimers versus heterodimers
- PMID: 36096675
- PMCID: PMC9468604
- DOI: 10.26508/lsa.202201474
Sibling rivalry among the ZBTB transcription factor family: homodimers versus heterodimers
Abstract
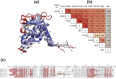
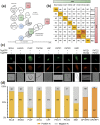
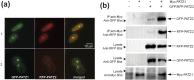
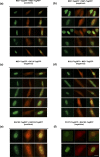
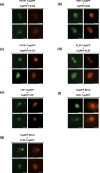
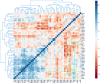
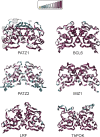
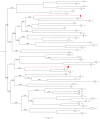
The BTB domain is an oligomerization domain found in over 300 proteins encoded in the human genome. In the family of BTB domain and zinc finger-containing (ZBTB) transcription factors, 49 members share the same protein architecture. The N-terminal BTB domain is structurally conserved among the family members and serves as the dimerization site, whereas the C-terminal zinc finger motifs mediate DNA binding. The available BTB domain structures from this family reveal a natural inclination for homodimerization. In this study, we investigated the potential for heterodimer formation in the cellular environment. We selected five BTB homodimers and four heterodimer structures. We performed cell-based binding assays with fluorescent protein-BTB domain fusions to assess dimer formation. We tested the binding of several BTB pairs, and we were able to confirm the heterodimeric physical interaction between the BTB domains of PATZ1 and PATZ2, previously reported only in an interactome mapping experiment. We also found this pair to be co-expressed in several immune system cell types. Finally, we used the available structures of BTB domain dimers and newly constructed models in extended molecular dynamics simulations (500 ns) to understand the energetic determinants of homo- and heterodimer formation. We conclude that heterodimer formation, although frequently described as less preferred than homodimers, is a possible mechanism to increase the combinatorial specificity of this transcription factor family.
© 2022 Piepoli et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous
