Genome-wide association analysis of 101 accessions dissects the genetic basis of shell thickness for genetic improvement in Persian walnut (Juglans regia L.)
- PMID: 36096735
- PMCID: PMC9469530
- DOI: 10.1186/s12870-022-03824-1
Genome-wide association analysis of 101 accessions dissects the genetic basis of shell thickness for genetic improvement in Persian walnut (Juglans regia L.)
Abstract
Background: Understanding the underlying genetic mechanisms that drive phenotypic variations is essential for enhancing the efficacy of crop improvement. Persian walnut (Juglans regia L.), which is grown extensively worldwide, is an important economic tree fruit due to its horticultural, medicinal, and material value. The quality of the walnut fruit is related to the selection of traits such as thinner shells, larger filling rates, and better taste, which is very important for breeding in China. The complex quantitative fruit-related traits are influenced by a variety of physiological and environmental factors, which can vary widely between walnut genotypes.
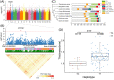
Results: For this study, a set of 101 Persian walnut accessions were re-sequenced, which generated a total of 906.2 Gb of Illumina sequence data with an average read depth of 13.8× for each accession. We performed the genome-wide association study (GWAS) using 10.9 Mb of high-quality single-nucleotide polymorphisms (SNPs) and 10 agronomic traits to explore the underlying genetic basis of the walnut fruit. Several candidate genes are proposed to be involved in walnut characteristics, including JrPXC1, JrWAKL8, JrGAMYB, and JrFRK1. Specifically, the JrPXC1 gene was confirmed to participate in the regulation of secondary wall cellulose thickening in the walnut shell.
Conclusion: In addition to providing considerable available genetic resources for walnut trees, this study revealed the underlying genetic basis involved in important walnut agronomic traits, particularly shell thickness, as well as providing clues for the improvement of genetic breeding and domestication in other perennial economic crops.
Keywords: Fruit-related traits; GWAS; Juglans regia; Shell thickness.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that this research was conducted in the absence of any commercial or financial relationships that might be construed as potential conflicts of interest.
Figures





Similar articles
-
Genome-wide patterns of population structure and association mapping of nut-related traits in Persian walnut populations from Iran using the Axiom J. regia 700K SNP array.Sci Rep. 2019 Apr 23;9(1):6376. doi: 10.1038/s41598-019-42940-1. Sci Rep. 2019. PMID: 31015545 Free PMC article.
-
Association and linkage mapping to unravel genetic architecture of phenological traits and lateral bearing in Persian walnut (Juglans regia L.).BMC Genomics. 2020 Mar 4;21(1):203. doi: 10.1186/s12864-020-6616-y. BMC Genomics. 2020. PMID: 32131731 Free PMC article.
-
Integrated multi-omics analyses provide new insights into genomic variation landscape and regulatory network candidate genes associated with walnut endocarp.Plant J. 2025 Apr;122(1):e70113. doi: 10.1111/tpj.70113. Plant J. 2025. PMID: 40162720
-
A Comprehensive Review on the Chemical Constituents and Functional Uses of Walnut (Juglans spp.) Husk.Int J Mol Sci. 2019 Aug 12;20(16):3920. doi: 10.3390/ijms20163920. Int J Mol Sci. 2019. PMID: 31409014 Free PMC article. Review.
-
Walnut (Juglans regia L.): genetic resources, chemistry, by-products.J Sci Food Agric. 2010 Sep;90(12):1959-67. doi: 10.1002/jsfa.4059. J Sci Food Agric. 2010. PMID: 20586084 Review.
Cited by
-
Complete mitochondrial genome assembly of Juglans regia unveiled its molecular characteristics, genome evolution, and phylogenetic implications.BMC Genomics. 2024 Sep 28;25(1):894. doi: 10.1186/s12864-024-10818-w. BMC Genomics. 2024. PMID: 39342114 Free PMC article.
-
Phenotypic Diversity Analysis and Integrative Evaluation of Camellia oleifera Germplasm Resources in Ya'an, Sichuan Province.Plants (Basel). 2025 Jul 21;14(14):2249. doi: 10.3390/plants14142249. Plants (Basel). 2025. PMID: 40733486 Free PMC article.
-
Genomic prediction in Persian walnut: Optimization levers according to genetic architecture of complex traits.Plant Genome. 2025 Jun;18(2):e70047. doi: 10.1002/tpg2.70047. Plant Genome. 2025. PMID: 40369746 Free PMC article.
References
-
- Zhao P, Zhou HJ, Potter D, Hu YH, Feng XJ, Dang M, Feng L, Zulfiqar S, Liu WZ, Zhao GF, et al. Population genetics, phylogenomics and hybrid speciation of Juglans in China determined from whole chloroplast genomes, transcriptomes, and genotyping-by-sequencing (GBS) Mol Phylogenet Evol. 2018;126:250–265. doi: 10.1016/j.ympev.2018.04.014. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

