Kinetic compartmentalization by unnatural reaction for itaconate production
- PMID: 36097012
- PMCID: PMC9468356
- DOI: 10.1038/s41467-022-33033-1
Kinetic compartmentalization by unnatural reaction for itaconate production
Abstract
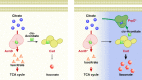
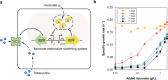
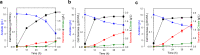
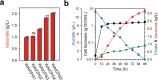
Physical compartmentalization of metabolism using membranous organelles in eukaryotes is helpful for chemical biosynthesis to ensure the availability of substrates from competitive metabolic reactions. Bacterial hosts lack such a membranous system, which is one of the major limitations for efficient metabolic engineering. Here, we employ kinetic compartmentalization with the introduction of an unnatural enzymatic reaction by an engineered enzyme as an alternative strategy to enable substrate availability from competitive reactions through kinetic isolation of metabolic pathways. As a proof of concept, we kinetically isolate the itaconate synthetic pathway from the tricarboxylic acid cycle in Escherichia coli, which is natively separated by mitochondrial membranes in Aspergillus terreus. Specifically, 2-methylcitrate dehydratase is engineered to alternatively catalyze citrate and kinetically secure cis-aconitate for efficient production using a high-throughput screening system. Itaconate production can be significantly improved with kinetic compartmentalization and its strategy has the potential to be widely applicable.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Cooper G. M. The Cell: A Molecular Approach (Sinauer Associates, 2000).
-
- Fani R. The origin and evolution of metabolic pathways: why and how did primordial cells construct metabolic routes? Evol. Educ. Outreach. 2012;5:367–381. doi: 10.1007/s12052-012-0439-5. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

