Genomic insights into rapid speciation within the world's largest tree genus Syzygium
- PMID: 36097018
- PMCID: PMC9468008
- DOI: 10.1038/s41467-022-32637-x
Genomic insights into rapid speciation within the world's largest tree genus Syzygium
Abstract
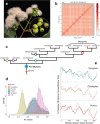
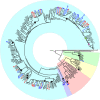
Species radiations, despite immense phenotypic variation, can be difficult to resolve phylogenetically when genetic change poorly matches the rapidity of diversification. Genomic potential furnished by palaeopolyploidy, and relative roles for adaptation, random drift and hybridisation in the apportionment of genetic variation, remain poorly understood factors. Here, we study these aspects in a model radiation, Syzygium, the most species-rich tree genus worldwide. Genomes of 182 distinct species and 58 unidentified taxa are compared against a chromosome-level reference genome of the sea apple, Syzygium grande. We show that while Syzygium shares an ancient genome doubling event with other Myrtales, little evidence exists for recent polyploidy events. Phylogenomics confirms that Syzygium originated in Australia-New Guinea and diversified in multiple migrations, eastward to the Pacific and westward to India and Africa, in bursts of speciation visible as poorly resolved branches on phylogenies. Furthermore, some sublineages demonstrate genomic clines that recapitulate cladogenetic events, suggesting that stepwise geographic speciation, a neutral process, has been important in Syzygium diversification.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Gittenberger E. What about non-adaptive radiation? Biol. J. Linn. Soc. 1991;43:263–272. doi: 10.1111/j.1095-8312.1991.tb00598.x. - DOI
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources

