Clonal hematopoiesis of indeterminate potential, DNA methylation, and risk for coronary artery disease
- PMID: 36097025
- PMCID: PMC9468335
- DOI: 10.1038/s41467-022-33093-3
Clonal hematopoiesis of indeterminate potential, DNA methylation, and risk for coronary artery disease
Abstract
Age-related changes to the genome-wide DNA methylation (DNAm) pattern observed in blood are well-documented. Clonal hematopoiesis of indeterminate potential (CHIP), characterized by the age-related acquisition and expansion of leukemogenic mutations in hematopoietic stem cells (HSCs), is associated with blood cancer and coronary artery disease (CAD). Epigenetic regulators DNMT3A and TET2 are the two most frequently mutated CHIP genes. Here, we present results from an epigenome-wide association study for CHIP in 582 Cardiovascular Health Study (CHS) participants, with replication in 2655 Atherosclerosis Risk in Communities (ARIC) Study participants. We show that DNMT3A and TET2 CHIP have distinct and directionally opposing genome-wide DNAm association patterns consistent with their regulatory roles, albeit both promoting self-renewal of HSCs. Mendelian randomization analyses indicate that a subset of DNAm alterations associated with these two leading CHIP genes may promote the risk for CAD.
© 2022. The Author(s).
Conflict of interest statement
B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. P.N. reports grant support from Amgen, Apple, AstraZeneca, Boston Scientific, and Novartis, spousal employment and equity at Vertex, consulting income from Apple, AstraZeneca, Novartis, Genentech / Roche, Blackstone Life Sciences, Foresite Labs, and TenSixteen Bio, and is a scientific advisor board member and shareholder of TenSixteen Bio and geneXwell, all unrelated to this work. J.S.F. has consulted for Shionogi Inc. R.B has consulted for Casana Care Inc, unrelated to this work. J.M. has guest-lectured at Merck, unrelated to this work. M.C.H. has consulted for CRISPR Therapeutics and served on the advisory board for Miga Health, both unrelated to this work. D.K. serves on a DSMB for Agnovos Healthcare, a scientific advisory board for Pfizer and Solarea Bio, and reports grant support from Amgen and Solarea Bio. B.L.E. has received research funding from Celgene, Deerfield, Novartis, and Calico and consulting fees from GRAIL, and is a member of the scientific advisory board and shareholder for Neomorph Inc., TenSixteen Bio, Skyhawk Therapeutics, and Exo Therapeutics. S.J. and A.G.B. are co-founders and equity holders in TenSixteen Bio. The other authors declare no competing interests.
Figures
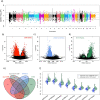
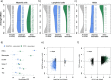

References
Publication types
MeSH terms
Grants and funding
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- RC2 HL102926/HL/NHLBI NIH HHS/United States
- N01 HC085082/HL/NHLBI NIH HHS/United States
- R01 HL148565/HL/NHLBI NIH HHS/United States
- R01 AR041398/AR/NIAMS NIH HHS/United States
- R01 HL142711/HL/NHLBI NIH HHS/United States
- DP2 HL157540/HL/NHLBI NIH HHS/United States
- RC2 HL103010/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- P30 ES009089/ES/NIEHS NIH HHS/United States
- R01 HL103612/HL/NHLBI NIH HHS/United States
- N01 HC085080/HL/NHLBI NIH HHS/United States
- DP5 OD029586/OD/NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- R01 HL151283/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- HHSN268201700004C/HB/NHLBI NIH HHS/United States
- R01 DK125782/DK/NIDDK NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- N01 HC085086/HL/NHLBI NIH HHS/United States
- R01 NS087541/NS/NINDS NIH HHS/United States
- N01 HC085083/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 HL086694/HL/NHLBI NIH HHS/United States
- T32 HG000040/HG/NHGRI NIH HHS/United States
- R01 HL087652/HL/NHLBI NIH HHS/United States
- R01 HL127564/HL/NHLBI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- K08 HL116640/HL/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- R01 HL146860/HL/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- HHSN268201700001I/HL/NHLBI NIH HHS/United States
- N01 HC055222/HL/NHLBI NIH HHS/United States
- R01 HL151152/HL/NHLBI NIH HHS/United States
- HHSN268201700004I/HL/NHLBI NIH HHS/United States
- N01 HC085079/HL/NHLBI NIH HHS/United States
- R01 AG059727/AG/NIA NIH HHS/United States
- U34 AG051418/AG/NIA NIH HHS/United States
- 75N92021D00006/HL/NHLBI NIH HHS/United States
- HHSN268201700005C/HL/NHLBI NIH HHS/United States
- HHSN268201700001C/HL/NHLBI NIH HHS/United States
- HHSN268201700002C/HB/NHLBI NIH HHS/United States
- HHSN268201700003C/HL/NHLBI NIH HHS/United States
- HHSN268201700002I/HL/NHLBI NIH HHS/United States
- HHSN268201700005I/HL/NHLBI NIH HHS/United States
- R01 HL135242/HL/NHLBI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- T32 AG047126/AG/NIA NIH HHS/United States
- HHSN268201700003I/HL/NHLBI NIH HHS/United States
- R01 HL148050/HL/NHLBI NIH HHS/United States
- N01 HC085081/HL/NHLBI NIH HHS/United States
- R01 HL141989/HL/NHLBI NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
- R01 HL111089/HL/NHLBI NIH HHS/United States
- R01 HL116747/HL/NHLBI NIH HHS/United States
- R01 HL092111/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

