Brain-enriched RagB isoforms regulate the dynamics of mTORC1 activity through GATOR1 inhibition
- PMID: 36097071
- PMCID: PMC9481464
- DOI: 10.1038/s41556-022-00977-x
Brain-enriched RagB isoforms regulate the dynamics of mTORC1 activity through GATOR1 inhibition
Abstract
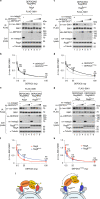
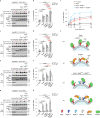
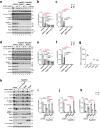
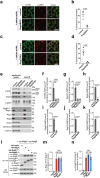
Mechanistic target of rapamycin complex 1 (mTORC1) senses nutrient availability to appropriately regulate cellular anabolism and catabolism. During nutrient restriction, different organs in an animal do not respond equally, with vital organs being relatively spared. This raises the possibility that mTORC1 is differentially regulated in different cell types, yet little is known about this mechanistically. The Rag GTPases, RagA or RagB bound to RagC or RagD, tether mTORC1 in a nutrient-dependent manner to lysosomes where mTORC1 becomes activated. Although the RagA and B paralogues were assumed to be functionally equivalent, we find here that the RagB isoforms, which are highly expressed in neurons, impart mTORC1 with resistance to nutrient starvation by inhibiting the RagA/B GTPase-activating protein GATOR1. We further show that high expression of RagB isoforms is observed in some tumours, revealing an alternative strategy by which cancer cells can retain elevated mTORC1 upon low nutrient availability.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





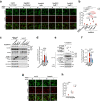
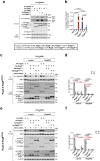
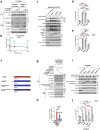
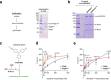


Comment in
-
Keeping up with the Rag GTPases.Nat Cell Biol. 2022 Sep;24(9):1330-1331. doi: 10.1038/s41556-022-00981-1. Nat Cell Biol. 2022. PMID: 36097073 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

