SREBP2-dependent lipid gene transcription enhances the infection of human dendritic cells by Zika virus
- PMID: 36097162
- PMCID: PMC9465152
- DOI: 10.1038/s41467-022-33041-1
SREBP2-dependent lipid gene transcription enhances the infection of human dendritic cells by Zika virus
Abstract
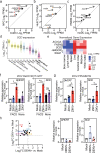
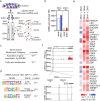
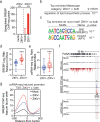
The emergence of Zika virus (ZIKV) as a global health threat has highlighted the unmet need for ZIKV-specific vaccines and antiviral treatments. ZIKV infects dendritic cells (DC), which have pivotal functions in activating innate and adaptive antiviral responses; however, the mechanisms by which DC function is subverted to establish ZIKV infection are unclear. Here we develop a genomics profiling method that enables discrete analysis of ZIKV-infected versus neighboring, uninfected primary human DCs to increase the sensitivity and specificity with which ZIKV-modulated pathways can be identified. The results show that ZIKV infection specifically increases the expression of genes enriched for lipid metabolism-related functions. ZIKV infection also increases the recruitment of sterol regulatory element-binding protein (SREBP) transcription factors to lipid gene promoters, while pharmacologic inhibition or genetic silencing of SREBP2 suppresses ZIKV infection of DCs. Our data thus identify SREBP2-activated transcription as a mechanism for promoting ZIKV infection amenable to therapeutic targeting.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA238662/CA/NCI NIH HHS/United States
- R01 NS106387/NS/NINDS NIH HHS/United States
- S10 OD026929/OD/NIH HHS/United States
- T32 GM145427/GM/NIGMS NIH HHS/United States
- R01 AI163188/AI/NIAID NIH HHS/United States
- R21 AI140063/AI/NIAID NIH HHS/United States
- R00 GM135515/GM/NIGMS NIH HHS/United States
- T32 GM008666/GM/NIGMS NIH HHS/United States
- R01 GM134366/GM/NIGMS NIH HHS/United States
- R01 NS103434/NS/NINDS NIH HHS/United States
- K99 GM135515/GM/NIGMS NIH HHS/United States
- R01 AI153500/AI/NIAID NIH HHS/United States
- K08 AI130381/AI/NIAID NIH HHS/United States
- R01 AI116813/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

