Independent phenotypic plasticity axes define distinct obesity sub-types
- PMID: 36097183
- PMCID: PMC9499872
- DOI: 10.1038/s42255-022-00629-2
Independent phenotypic plasticity axes define distinct obesity sub-types
Abstract
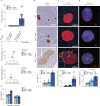
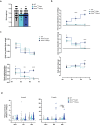
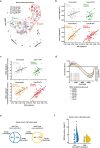
Studies in genetically 'identical' individuals indicate that as much as 50% of complex trait variation cannot be traced to genetics or to the environment. The mechanisms that generate this 'unexplained' phenotypic variation (UPV) remain largely unknown. Here, we identify neuronatin (NNAT) as a conserved factor that buffers against UPV. We find that Nnat deficiency in isogenic mice triggers the emergence of a bi-stable polyphenism, where littermates emerge into adulthood either 'normal' or 'overgrown'. Mechanistically, this is mediated by an insulin-dependent overgrowth that arises from histone deacetylase (HDAC)-dependent β-cell hyperproliferation. A multi-dimensional analysis of monozygotic twin discordance reveals the existence of two patterns of human UPV, one of which (Type B) phenocopies the NNAT-buffered polyphenism identified in mice. Specifically, Type-B monozygotic co-twins exhibit coordinated increases in fat and lean mass across the body; decreased NNAT expression; increased HDAC-responsive gene signatures; and clinical outcomes linked to insulinemia. Critically, the Type-B UPV signature stratifies both childhood and adult cohorts into four metabolic states, including two phenotypically and molecularly distinct types of obesity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











Comment in
-
A stochastic basis for metabolic phenotypes.Nat Metab. 2022 Sep;4(9):1091-1092. doi: 10.1038/s42255-022-00630-9. Nat Metab. 2022. PMID: 36097184 No abstract available.
-
Obesity phenotypes: explaining the unexplained.Nat Rev Endocrinol. 2022 Dec;18(12):717. doi: 10.1038/s41574-022-00760-w. Nat Rev Endocrinol. 2022. PMID: 36207625 No abstract available.
References
-
- Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edin. 1918;52:399–433. doi: 10.1017/S0080456800012163. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

