RNF43 mutations predict response to anti-BRAF/EGFR combinatory therapies in BRAFV600E metastatic colorectal cancer
- PMID: 36097219
- PMCID: PMC9556333
- DOI: 10.1038/s41591-022-01976-z
RNF43 mutations predict response to anti-BRAF/EGFR combinatory therapies in BRAFV600E metastatic colorectal cancer
Abstract
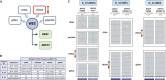
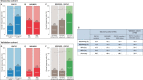
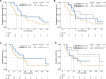
Anti-BRAF/EGFR therapy was recently approved for the treatment of metastatic BRAFV600E colorectal cancer (mCRCBRAF-V600E). However, a large fraction of patients do not respond, underscoring the need to identify molecular determinants of treatment response. Using whole-exome sequencing in a discovery cohort of patients with mCRCBRAF-V600E treated with anti-BRAF/EGFR therapy, we found that inactivating mutations in RNF43, a negative regulator of WNT, predict improved response rates and survival outcomes in patients with microsatellite-stable (MSS) tumors. Analysis of an independent validation cohort confirmed the relevance of RNF43 mutations to predicting clinical benefit (72.7% versus 30.8%; P = 0.03), as well as longer progression-free survival (hazard ratio (HR), 0.30; 95% confidence interval (CI), 0.12-0.75; P = 0.01) and overall survival (HR, 0.26; 95% CI, 0.10-0.71; P = 0.008), in patients with MSS-RNF43mutated versus MSS-RNF43wild-type tumors. Microsatellite-instable tumors invariably carried a wild-type-like RNF43 genotype encoding p.G659fs and presented an intermediate response profile. We found no association of RNF43 mutations with patient outcomes in a control cohort of patients with MSS-mCRCBRAF-V600E tumors not exposed to anti-BRAF targeted therapies. Overall, our findings suggest a cross-talk between the MAPK and WNT pathways that may modulate the antitumor activity of anti-BRAF/EGFR therapy and uncover predictive biomarkers to optimize the clinical management of these patients.
© 2022. The Author(s).
Conflict of interest statement
C.C. reports receiving honoraria and speaker’s bureau compensation from Servier, Bayer, Merck, Amgen, Pierre Fabre, MSD, Roche and Nordic Pharma and research grants from Merck, Servier and Amgen. D.G. was an employee of Novartis and was a shareholder while this work was being conducted. D.P.K. is an employee of Novartis. E.E. reports receiving honoraria for an advisory role, travel grants and research grants (past 5 years) from Hoffmann-La Roche, Bristol-Myers Squibb, Servier, Amgen, Merck Serono, Array Biopharma, Sanofi and Bayer. Her institution received honoraria due to her investigator contribution in clinical trials from Array Biopharma, MSD, Abbvie, Amgen, GlaxoSmithKline, AstraZeneca, Merck Sharp & Dohme, Bristol-Myers Squibb, Novartis, Boehringer Ingelheim, Hoffmann-La Roche, Medimmune, Pierre-Fabre and Sanofi Aventis. F.P. received honoraria from Amgen, Merck Serono, Lilly, Sanofi, Bayer, Servier, MSD, AstraZeneca and Organon and research grants from AstraZeneca and BMS. G.K. is an employee of Novartis. G.V. has received a speaker’s fee from MSD and Pierre Fabrer and has held an advisory role with AstraZeneca. J.R. declares receiving honoraria from Sanofi and travel and accommodation expenses from Amgen, Merck and Sanofi. J.T. reports having a personal financial interest in the form of a scientific consultancy role for Array Biopharma, AstraZeneca, Avvinity, Bayer, Boehringer Ingelheim, Chugai, Daiichi Sankyo, F. Hoffmann-La Roche, Genentech, HalioDX, Hutchison MediPharma International, Ikena Oncology, Inspirna, IQVIA, Lilly, Menarini, Merck Serono, Merus, MSD, Mirati, Neophore, Novartis, Ona Therapeutics, Orion Biotechnology, Peptomyc, Pfizer, Pierre Fabre, Samsung Bioepis, Sanofi, Seattle Genetics, Scandion Oncology, Servier, Sotio Biotech, Taiho, Tessa Therapeutics and TheraMyc, as well as educational collaboration with Imedex, Medscape Education, MJH Life Sciences, PeerView Institute for Medical Education and Physicians Education Resource (PER). P.N. reports receiving honoraria or consultation fees from Novartis, Bayer and MSD Oncology and had travel and accommodation paid for or reimbursed by Novartis. R.A.T. reports receiving a research grant related to this study from Novartis and research grants unrelated to this study from AstraZeneca and Beigene. R.D. declares an advisory role for Roche and Boehringer Ingelheim and received a speaker’s fee from Roche, Boehringer Ingelheim, Ipsen, Amgen, Servier, Sanofi, Libbs and Merck Sharp & Dohme and research grants from Merck and Pierre Fabre. R.P.-L. has acted in a consulting or advisory role for Roche, and she has participated in speaker bureaus sponsored by Roche and Pfizer. She is also the principal investigator in research grants to VHIO funded by AstraZeneca and Roche. S. Lonardi declares no conflict of interest inside the scope of the present work but declares interests in other projects from a consulting or advisory role (Amgen, Merck Serono, Lilly, AstraZeneca, Incyte, Daiichi-Sankyo, Bristol-Myers Squibb, Servier and MSD); speaker’s bureau compensation from Roche, Lilly, Bristol-Myers Squibb, Servier, Merck Serono, Pierre-Fabre, GSK and Amgen; and research funding from Amgen, Merck Serono, Bayer, Roche, Lilly, AstraZeneca and Bristol-Myers Squibb. The other authors declare no conflicts of interest.
Figures
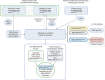

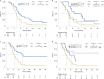

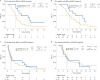


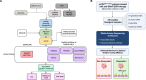




References
-
- Cremolini C, et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015;16:1306–1315. doi: 10.1016/S1470-2045(15)00122-9. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

