On the impermanence of species: The collapse of genetic incompatibilities in hybridizing populations
- PMID: 36097352
- PMCID: PMC9827863
- DOI: 10.1111/evo.14626
On the impermanence of species: The collapse of genetic incompatibilities in hybridizing populations
Abstract
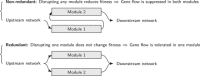
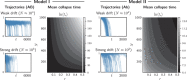
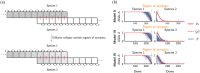
Species pairs often become genetically incompatible during divergence, which is an important source of reproductive isolation. An idealized picture is often painted where incompatibility alleles accumulate and fix between diverging species. However, recent studies have shown both that incompatibilities can collapse with ongoing hybridization, and that incompatibility loci can be polymorphic within species. This paper suggests some general rules for the behavior of incompatibilities under hybridization. In particular, we argue that redundancy of genetic pathways can strongly affect the dynamics of intrinsic incompatibilities. Since fitness in genetically redundant systems is unaffected by introducing a few foreign alleles, higher redundancy decreases the stability of incompatibilities during hybridization, but also increases tolerance of incompatibility polymorphism within species. We use simulations and theories to show that this principle leads to two types of collapse: in redundant systems, exemplified by classical Dobzhansky-Muller incompatibilities, collapse is continuous and approaches a quasi-neutral polymorphism between broadly sympatric species, often as a result of isolation-by-distance. In nonredundant systems, exemplified by co-evolution among genetic elements, incompatibilities are often stable, but can collapse abruptly with spatial traveling waves. As both types are common, the proposed principle may be useful in understanding the abundance of genetic incompatibilities in natural populations.
Keywords: Genetic redundancy; hybridization; incompatibility; purging of incompatibilities; reproductive isolation.
© 2022 The Authors. Evolution published by Wiley Periodicals LLC on behalf of The Society for the Study of Evolution.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Comment in
-
Digest: The non-redundancy of non-ephemeral reproductive isolation.Evolution. 2023 Feb 4;77(2):618-619. doi: 10.1093/evolut/qpac060. Evolution. 2023. PMID: 36622336
References
-
- Atlan, A. , Capillon, C. , Derome, N. , Couvet, D. & Montchamp‐Moreau, C. (2003) The evolution of autosomal suppressors of sex‐ratio drive in Drosophila simulans . Genetica, 117(1), 47–58. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
