Bio-SODA UX: enabling natural language question answering over knowledge graphs with user disambiguation
- PMID: 36097541
- PMCID: PMC9458692
- DOI: 10.1007/s10619-022-07414-w
Bio-SODA UX: enabling natural language question answering over knowledge graphs with user disambiguation
Abstract
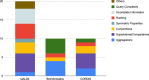
The problem of natural language processing over structured data has become a growing research field, both within the relational database and the Semantic Web community, with significant efforts involved in question answering over knowledge graphs (KGQA). However, many of these approaches are either specifically targeted at open-domain question answering using DBpedia, or require large training datasets to translate a natural language question to SPARQL in order to query the knowledge graph. Hence, these approaches often cannot be applied directly to complex scientific datasets where no prior training data is available. In this paper, we focus on the challenges of natural language processing over knowledge graphs of scientific datasets. In particular, we introduce Bio-SODA, a natural language processing engine that does not require training data in the form of question-answer pairs for generating SPARQL queries. Bio-SODA uses a generic graph-based approach for translating user questions to a ranked list of SPARQL candidate queries. Furthermore, Bio-SODA uses a novel ranking algorithm that includes node centrality as a measure of relevance for selecting the best SPARQL candidate query. Our experiments with real-world datasets across several scientific domains, including the official bioinformatics Question Answering over Linked Data (QALD) challenge, as well as the CORDIS dataset of European projects, show that Bio-SODA outperforms publicly available KGQA systems by an F1-score of least 20% and by an even higher factor on more complex bioinformatics datasets. Finally, we introduce Bio-SODA UX, a graphical user interface designed to assist users in the exploration of large knowledge graphs and in dynamically disambiguating natural language questions that target the data available in these graphs.
Keywords: Knowledge graphs; Question answering; Ranking.
© The Author(s) 2022.
Figures



References
-
- Diefenbach D, Both A, Singh K, Maret P. Towards a question answering system over the semantic web. Semantic Web Preprint. 2018;2018:1–19.
-
- Zheng, W., Yu, J.X., Zou, L., Cheng, H.: Question answering over knowledge graphs: question understanding via template decomposition. In: Proceedings of the VLDB Endowment 11, pp. 1373–1386 (2018)
-
- Vakulenko, S., Garcia, J.D.F., Polleres, A., de Rijke, M., Cochez, M.: Message Passing for Complex Question Answering over Knowledge Graphs. In: Proceedings of the 28th ACM International Conference on Information and Knowledge Management, pp. 1431–1440 (2019)
-
- Li F, Jagadish HV. Constructing an interactive natural language interface for relational databases. Proc. VLDB Endowm. 2014;8:73–84. doi: 10.14778/2735461.2735468. - DOI
-
- Li F, Jagadish HV. Understanding natural language queries over relational databases. ACM SIGMOD Rec. 2016;45:6–13. doi: 10.1145/2949741.2949744. - DOI
LinkOut - more resources
Full Text Sources
