Enhancement of catalytic activity and alkaline stability of cellobiohydrolase by structure-based protein engineering
- PMID: 36097631
- PMCID: PMC9463429
- DOI: 10.1007/s13205-022-03339-4
Enhancement of catalytic activity and alkaline stability of cellobiohydrolase by structure-based protein engineering
Abstract
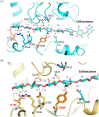
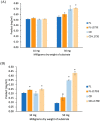
Alkaline cellobiohydrolases have the potential for application in various industries, including pulp processing and laundry where operation under high pH conditions is preferred. In this study, variants of CtCel6A cellobiohydrolase from Chaetomium thermophilum were generated by structural-based protein engineering with the rationale of increasing catalytic activity and alkaline stability. The variants included removal of the carbohydrate-binding module (CBM) and substitution of residues 173 and 200. The CBM-deleted enzyme with Y200F mutation predicted to mediate conformational change at the N-terminal loop demonstrated increased alkaline stability at 60 °C, pH 8.0 for 24 h up to 2.25-fold compared with the wild-type enzyme. Another CBM-deleted enzyme with L173E mutation predicted to induce a new hydrogen bond in the substrate-binding cleft showed enhanced hydrolysis yield of pretreated sugarcane trash up to 4.65-fold greater than that of the wild-type enzyme at the pH 8.0. The variant enzymes could thus be developed for applications on cellulose hydrolysis and plant fiber modification operated under alkaline conditions.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-022-03339-4.
Keywords: Alkaline stability; Cellobiohydrolase; Pulp and paper; Structure-based protein engineering.
© King Abdulaziz City for Science and Technology 2022, Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures



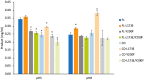
References
-
- Baramee S, Teeravivattanakit T, Phitsuwan P, Waeonukul R, Pason P, Tachaapaikoon C, Kosugi A, Sakka K, Ratanakhanokchai K. A novel GH6 cellobiohydrolase from Paenibacillus curdlanolyticus B-6 and its synergistic action on cellulose degradation. Appl Microbiol Biotechnol. 2017;101(3):1175–1188. doi: 10.1007/s00253-016-7895-8. - DOI - PubMed
-
- Becker D, Braet C, Brumer H, Claeyssens M, Divne C, Fagerström R, Harris M, Jones T, Kleywegt G, Koivula A, Mahdi S, Piens K, Sinnott M, Ståhlberg J, Teeri T, Underwood M, Wohlfahrt G. Engineering of a glycosidase Family 7 cellobiohydrolase to more alkaline pH optimum: the pH behaviour of Trichoderma reesei CeI7A and its E223S/A224H/L225V/T226A/D262G mutant. Biochem J. 2001;356:19–30. doi: 10.1042/0264-6021:3560019. - DOI - PMC - PubMed
-
- Bernardes A, Pellegrini VOA, Curtolo F, Camilo CM, Mello BL, Johns MA, Scott JL, Guimaraes FEC, Polikarpov I. Carbohydrate binding modules enhance cellulose enzymatic hydrolysis by increasing access of cellulases to the substrate. Carbohydr Polym. 2019;211:57–68. doi: 10.1016/j.carbpol.2019.01.108. - DOI - PubMed
LinkOut - more resources
Full Text Sources

