Reduced and Nonreduced Genomes in Paraburkholderia Symbionts of Social Amoebas
- PMID: 36098425
- PMCID: PMC9601139
- DOI: 10.1128/msystems.00562-22
Reduced and Nonreduced Genomes in Paraburkholderia Symbionts of Social Amoebas
Erratum in
-
Correction for Noh et al., "Reduced and Nonreduced Genomes in Paraburkholderia Symbionts of Social Amoebas".mSystems. 2024 Oct 22;9(10):e0106524. doi: 10.1128/msystems.01065-24. Epub 2024 Sep 18. mSystems. 2024. PMID: 39291992 Free PMC article. No abstract available.
Abstract
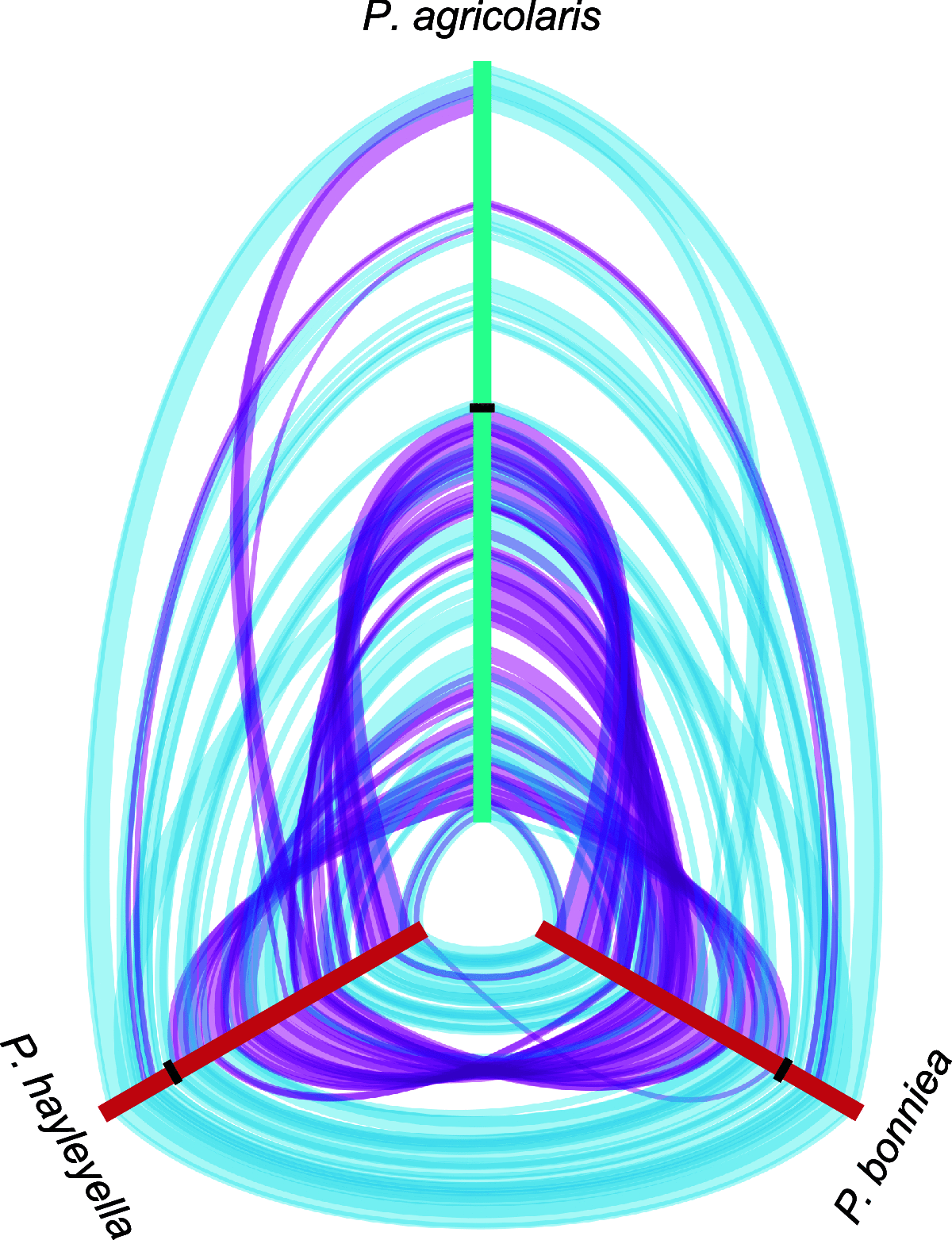
The social amoeba Dictyostelium discoideum is a predatory soil protist frequently used for studying host-pathogen interactions. A subset of D. discoideum strains isolated from soil persistently carry symbiotic Paraburkholderia, recently formally described as P. agricolaris, P. bonniea, and P. hayleyella. The three facultative symbiont species of D. discoideum present a unique opportunity to study a naturally occurring symbiosis in a laboratory model protist. There is a large difference in genome size between P. agricolaris (8.7 million base pairs [Mbp]) versus P. hayleyella and P. bonniea (4.1 Mbp). We took a comparative genomics approach and compared the three genomes of D. discoideum symbionts to 12 additional Paraburkholderia genomes to test for genome evolution patterns that frequently accompany host adaptation. Overall, P. agricolaris is difficult to distinguish from other Paraburkholderia based on its genome size and content, but the reduced genomes of P. bonniea and P. hayleyella display characteristics indicative of genome streamlining rather than deterioration during adaptation to their protist hosts. In addition, D. discoideum-symbiont genomes have increased secretion system and motility genes that may mediate interactions with their host. Specifically, adjacent BurBor-like type 3 and T6SS-5-like type 6 secretion system operons shared among all three D. discoideum-symbiont genomes may be important for host interaction. Horizontal transfer of these secretion system operons within the amoeba host environment may have contributed to the unique ability of these symbionts to establish and maintain a symbiotic relationship with D. discoideum. IMPORTANCE Protists are a diverse group of typically single cell eukaryotes. Bacteria and archaea that form long-term symbiotic relationships with protists may evolve in additional ways than those in relationships with multicellular eukaryotes such as plants, animals, or fungi. Social amoebas are a predatory soil protist sometimes found with symbiotic bacteria living inside their cells. They present a unique opportunity to explore a naturally occurring symbiosis in a protist frequently used for studying host-pathogen interactions. We show that one amoeba-symbiont species is similar to other related bacteria in genome size and content, while the two reduced-genome-symbiont species show characteristics of genome streamlining rather than deterioration during adaptation to their host. We also identify sets of genes present in all three amoeba-symbiont genomes that are potentially used for host-symbiont interactions. Because the amoeba symbionts are distantly related, the amoeba host environment may be where these genes were shared among symbionts.
Keywords: Burkholderia; Dictyostelium; genome reduction; protist; symbiosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

