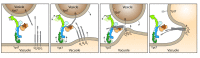
Structure of the HOPS tethering complex, a lysosomal membrane fusion machinery
- PMID: 36098503
- PMCID: PMC9592082
- DOI: 10.7554/eLife.80901
Structure of the HOPS tethering complex, a lysosomal membrane fusion machinery
Abstract
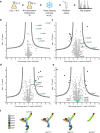
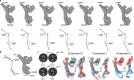
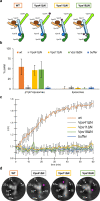
Lysosomes are essential for cellular recycling, nutrient signaling, autophagy, and pathogenic bacteria and viruses invasion. Lysosomal fusion is fundamental to cell survival and requires HOPS, a conserved heterohexameric tethering complex. On the membranes to be fused, HOPS binds small membrane-associated GTPases and assembles SNAREs for fusion, but how the complex fulfills its function remained speculative. Here, we used cryo-electron microscopy to reveal the structure of HOPS. Unlike previously reported, significant flexibility of HOPS is confined to its extremities, where GTPase binding occurs. The SNARE-binding module is firmly attached to the core, therefore, ideally positioned between the membranes to catalyze fusion. Our data suggest a model for how HOPS fulfills its dual functionality of tethering and fusion and indicate why it is an essential part of the membrane fusion machinery.
Keywords: HOPS; Rab GTPase; S. cerevisiae; cell biology; lysosome; membrane fusion; molecular biophysics; structural biology; tethering; vacuole.
Plain language summary
Our cells break down the nutrients that they receive from the body to create the building blocks needed to keep us alive. This is done by compartments called lysosomes that are filled with a cocktail of proteins called enzymes, which speed up the breakdown process. Lysosomes are surrounded by a membrane, a barrier of fatty molecules that protects the rest of the cell from being digested. When new nutrients reach the cell, they travel to the lysosome packaged in vesicles, which have their own fatty membrane. To allow the nutrients to enter the lysosome without creating a leak, the membranes of the vesicles and the lysosome must fuse. The mechanism through which these membranes fuse is not fully clear. It is known that both fusing membranes must contain proteins called SNAREs, which wind around each other when they interact. However, this alone is not enough. Other proteins are also required to tether the membranes together before they fuse. To understand how these tethers play a role, Shvarev, Schoppe, König et al. studied the structure of the HOPS complex from yeast. This assembly of six proteins is vital for lysosomal fusion and, has a composition similar to the equivalent complex in humans. Using cryo-electron microscopy, a technique that relies on freezing purified proteins to image them with an electron microscope and reveal their structure, allowed Shvarev, Schoppe, König et al. to provide a model for how HOPS interacts with SNAREs and membranes. In addition to HOPS acting as a tether to bring the membranes together, it can also bind directly to SNAREs. This creates a bridge that allows the proteins to wrap around each other, driving the membranes to fuse. HOPS is a crucial component in the cellular machinery, and mutations in the complex can cause devastating neurological defects. The complex is also targeted by viruses – such as SARS-CoV-2 – that manipulate HOPS to reduce its activity. Shvarev, Schoppe, König et al.’s findings could help researchers to develop drugs to maintain or recover the activity of HOPS. However, this will require additional information about its structure and how the complex acts in the biological environment of the cell.
© 2022, Shvarev, Schoppe, König et al.
Conflict of interest statement
DS, JS, CK, AP, NF, SK, LL, DJ, KS, DK, FF, AM, CU No competing interests declared
Figures


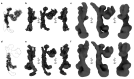







References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica. Section D, Biological Crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

