Structural aspects of hepatitis E virus
- PMID: 36098802
- PMCID: PMC9469829
- DOI: 10.1007/s00705-022-05575-8
Structural aspects of hepatitis E virus
Abstract
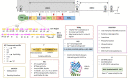
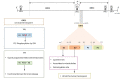
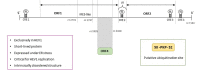
Hepatitis E virus (HEV) is a leading cause of acute hepatitis worldwide. Hepatitis E is an enterically transmitted zoonotic disease that causes large waterborne epidemic outbreaks in developing countries and has become an increasing public-health concern in industrialized countries. In this setting, the infection is usually acute and self-limiting in immunocompetent individuals, although chronic cases in immunocompromised patients have been reported, frequently associated with several extrahepatic manifestations. Moreover, extrahepatic manifestations have also been reported in immunocompetent individuals with acute HEV infection. HEV belongs to the alphavirus-like supergroup III of single-stranded positive-sense RNA viruses, and its genome contains three partially overlapping open reading frames (ORFs). ORF1 encodes a nonstructural protein with eight domains, most of which have not been extensively characterized: methyltransferase, Y domain, papain-like cysteine protease, hypervariable region, proline-rich region, X domain, Hel domain, and RNA-dependent RNA polymerase. ORF2 and ORF3 encode the capsid protein and a multifunctional protein believed to be involved in virion release, respectively. The novel ORF4 is only expressed in HEV genotype 1 under endoplasmic reticulum stress conditions, and its exact function has not yet been elucidated. Despite important advances in recent years, the biological and molecular processes underlying HEV replication remain poorly understood, primarily due to a lack of detailed information about the functions of the viral proteins and the mechanisms involved in host-pathogen interactions. This review summarizes the current knowledge concerning HEV proteins and their biological properties, providing updated detailed data describing their function and focusing in detail on their structural characteristics. Furthermore, we review some unclear aspects of the four proteins encoded by the ORFs, highlighting the current key information gaps and discussing potential novel experimental strategies for shedding light on those issues.
Keywords: Hepatitis E virus; Review; Structural biology.
© 2022. The Author(s), under exclusive licence to Springer-Verlag GmbH Austria, part of Springer Nature.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
-
- Anang S, Subramani C, Nair VP, Kaul S, Kaushik N, Sharma C, Tiwari A, Ranjith-Kumar CT, Surjit M. Identification of critical residues in Hepatitis E virus macro domain involved in its interaction with viral methyltransferase and ORF3 proteins. Sci Rep. 2016;6:25133. doi: 10.1038/srep25133. - DOI - PMC - PubMed
-
- Ansari IH, Nanda SK, Durgapal H, Agrawal S, Mohanty SK, Gupta D, Jameel S, Panda SK. Cloning, sequencing, and expression of the hepatitis E virus (HEV) nonstructural open reading frame 1 (ORF1) J Med Virol. 2000;60:275–283. doi: 10.1002/(SICI)1096-9071(200003)60:3<275::AID-JMV5>3.0.CO;2-9. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

