Single-cell transcriptomics reveals skewed cellular communication and phenotypic shift in pulmonary artery remodeling
- PMID: 36099047
- PMCID: PMC9714792
- DOI: 10.1172/jci.insight.153471
Single-cell transcriptomics reveals skewed cellular communication and phenotypic shift in pulmonary artery remodeling
Abstract
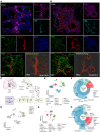
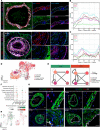
A central feature of progressive vascular remodeling is altered smooth muscle cell (SMC) homeostasis; however, the understanding of how different cell populations contribute to this process is limited. Here, we utilized single-cell RNA sequencing to provide insight into cellular composition changes within isolated pulmonary arteries (PAs) from pulmonary arterial hypertension and donor lungs. Our results revealed that remodeling skewed the balanced communication network between immune and structural cells, in particular SMCs. Comparative analysis with murine PAs showed that human PAs harbored heterogeneous SMC populations with an abundant intermediary cluster displaying a gradient transition between SMCs and adventitial fibroblasts. Transcriptionally distinct SMC populations were enriched in specific biological processes and could be differentiated into 4 major clusters: oxygen sensing (enriched in pericytes), contractile, synthetic, and fibroblast-like. End-stage remodeling was associated with phenotypic shift of preexisting SMC populations and accumulation of synthetic SMCs in neointima. Distinctly regulated genes in clusters built nonredundant regulatory hubs encompassing stress response and differentiation regulators. The current study provides a blueprint of cellular and molecular changes on a single-cell level that are defining the pathological vascular remodeling process.
Keywords: Hypertension; Pulmonology; Vascular Biology.
Figures





References
-
- Hoffmann J, et al. Distinct differences in gene expression patterns in pulmonary arteries of patients with chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis with pulmonary hypertension. Am J Respir Crit Care Med. 2014;190(1):98–111. doi: 10.1164/rccm.201401-0037OC. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL140409/HL/NHLBI NIH HHS/United States
- R01 HL087825/HL/NHLBI NIH HHS/United States
- R01 HL164014/HL/NHLBI NIH HHS/United States
- R01 HL148810/HL/NHLBI NIH HHS/United States
- K08 HL150226/HL/NHLBI NIH HHS/United States
- R01 HL132349/HL/NHLBI NIH HHS/United States
- U01 HL134745/HL/NHLBI NIH HHS/United States
- K08 HL163398/HL/NHLBI NIH HHS/United States
- U01 HL110942/HL/NHLBI NIH HHS/United States
- R01 HL162299/HL/NHLBI NIH HHS/United States
- R01 HL132999/HL/NHLBI NIH HHS/United States
- R01 HL133951/HL/NHLBI NIH HHS/United States
- R01 HL123957/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

