Immune dysfunction signatures predict outcomes and define checkpoint blockade-unresponsive microenvironments in acute myeloid leukemia
- PMID: 36099049
- PMCID: PMC9621145
- DOI: 10.1172/JCI159579
Immune dysfunction signatures predict outcomes and define checkpoint blockade-unresponsive microenvironments in acute myeloid leukemia
Abstract
BackgroundImmune exhaustion and senescence are dominant dysfunctional states of effector T cells and major hurdles for the success of cancer immunotherapy. In the current study, we characterized how acute myeloid leukemia (AML) promotes the generation of senescent-like CD8+ T cells and whether they have prognostic relevance.METHODSWe analyzed NanoString, bulk RNA-Seq and single-cell RNA-Seq data from independent clinical cohorts comprising 1,896 patients treated with chemotherapy and/or immune checkpoint blockade (ICB).ResultsWe show that senescent-like bone marrow CD8+ T cells were impaired in killing autologous AML blasts and that their proportion negatively correlated with overall survival (OS). We defined what we believe to be new immune effector dysfunction (IED) signatures using 2 gene expression profiling platforms and reported that IED scores correlated with adverse-risk molecular lesions, stemness, and poor outcomes; these scores were a more powerful predictor of OS than 2017-ELN risk or leukemia stem cell (LSC17) scores. IED expression signatures also identified an ICB-unresponsive tumor microenvironment and predicted significantly shorter OS.ConclusionThe IED scores provided improved AML-risk stratification and could facilitate the delivery of personalized immunotherapies to patients who are most likely to benefit.TRIAL REGISTRATIONClinicalTrials.gov; NCT02845297.FUNDINGJohn and Lucille van Geest Foundation, Nottingham Trent University's Health & Wellbeing Strategic Research Theme, NIH/NCI P01CA225618, Genentech-imCORE ML40354, Qatar National Research Fund (NPRP8-2297-3-494).
Keywords: Cancer immunotherapy; Cellular senescence; Hematology; Leukemias.
Figures
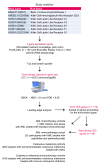
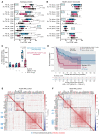
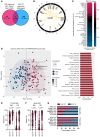
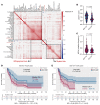
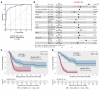
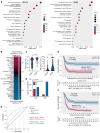
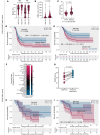

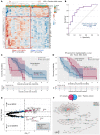

Comment in
-
Integrated analysis of single-cell RNA-seq and bulk RNA-seq reveals RNA N6-methyladenosine modification associated with prognosis and drug resistance in acute myeloid leukemia.Front Immunol. 2023 Oct 31;14:1281687. doi: 10.3389/fimmu.2023.1281687. eCollection 2023. Front Immunol. 2023. PMID: 38022588 Free PMC article.
References
-
- Daver N, et al. Efficacy, safety, and biomarkers of response to azacitidine and nivolumab in relapsed/refractory acute myeloid leukemia: a nonrandomized, open-label, phase II study. Cancer Discov. 2019;9(3):370–383. doi: 10.1158/2159-8290.CD-18-0774. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

