Lamin A/C impairments cause mitochondrial dysfunction by attenuating PGC1α and the NAMPT-NAD+ pathway
- PMID: 36099415
- PMCID: PMC9508839
- DOI: 10.1093/nar/gkac741
Lamin A/C impairments cause mitochondrial dysfunction by attenuating PGC1α and the NAMPT-NAD+ pathway
Abstract
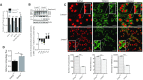
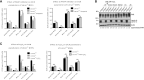
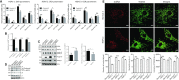
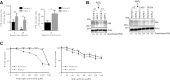
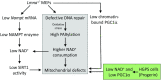
Mutations in the lamin A/C gene (LMNA) cause laminopathies such as the premature aging Hutchinson Gilford progeria syndrome (HGPS) and altered lamin A/C levels are found in diverse malignancies. The underlying lamin-associated mechanisms remain poorly understood. Here we report that lamin A/C-null mouse embryo fibroblasts (Lmna-/- MEFs) and human progerin-expressing HGPS fibroblasts both display reduced NAD+ levels, unstable mitochondrial DNA and attenuated bioenergetics. This mitochondrial dysfunction is associated with reduced chromatin recruitment (Lmna-/- MEFs) or low levels (HGPS) of PGC1α, the key transcription factor for mitochondrial homeostasis. Lmna-/- MEFs showed reduced expression of the NAD+-biosynthesis enzyme NAMPT and attenuated activity of the NAD+-dependent deacetylase SIRT1. We find high PARylation in lamin A/C-aberrant cells, further decreasing the NAD+ pool and consistent with impaired DNA base excision repair in both cell models, a condition that fuels DNA damage-induced PARylation under oxidative stress. Further, ATAC-sequencing revealed a substantially altered chromatin landscape in Lmna-/- MEFs, including aberrantly reduced accessibility at the Nampt gene promoter. Thus, we identified a new role of lamin A/C as a key modulator of mitochondrial function through impairments of PGC1α and the NAMPT-NAD+ pathway, with broader implications for the aging process.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Burke B., Stewart C.L.. The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol. 2013; 14:13–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

