A toolkit for the identification of NEAT1_2/paraspeckle modulators
- PMID: 36099417
- PMCID: PMC9723620
- DOI: 10.1093/nar/gkac771
A toolkit for the identification of NEAT1_2/paraspeckle modulators
Abstract
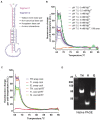
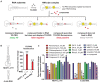
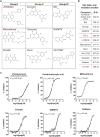
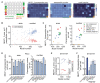
Paraspeckles are ribonucleoprotein granules assembled by NEAT1_2 lncRNA, an isoform of Nuclear Paraspeckle Assembly Transcript 1 (NEAT1). Dysregulation of NEAT1_2/paraspeckles has been linked to multiple human diseases making them an attractive drug target. However currently NEAT1_2/paraspeckle-focused translational research and drug discovery are hindered by a limited toolkit. To fill this gap, we developed and validated a set of tools for the identification of NEAT1_2 binders and modulators comprised of biochemical and cell-based assays. The NEAT1_2 triple helix stability element was utilized as the target in the biochemical assays, and the cellular assay ('ParaQuant') was based on high-content imaging of NEAT1_2 in fixed cells. As a proof of principle, these assays were used to screen a 1,200-compound FDA-approved drug library and a 170-compound kinase inhibitor library and to confirm the screening hits. The assays are simple to establish, use only commercially-available reagents and are scalable for higher throughput. In particular, ParaQuant is a cost-efficient assay suitable for any cells growing in adherent culture and amenable to multiplexing. Using ParaQuant, we identified dual PI3K/mTOR inhibitors as potent negative modulators of paraspeckles. The tools we describe herein should boost paraspeckle studies and help guide the search, validation and optimization of NEAT1_2/paraspeckle-targeted small molecules.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Fox A.H., Nakagawa S., Hirose T., Bond C.S.. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 2018; 43:124–135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

