Tutorial: Microbiome studies in drug metabolism
- PMID: 36099474
- PMCID: PMC9747132
- DOI: 10.1111/cts.13416
Tutorial: Microbiome studies in drug metabolism
Abstract
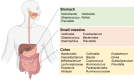
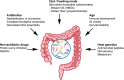
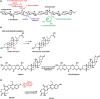
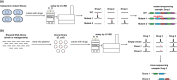
The human gastrointestinal tract is home to a dense population of microorganisms whose metabolism impacts human health and physiology. The gut microbiome encodes millions of genes, the products of which endow our bodies with unique biochemical activities. In the context of drug metabolism, microbial biochemistry in the gut influences humans in two major ways: (1) by producing small molecules that modulate expression and activity of human phase I and II pathways; and (2) by directly modifying drugs administered to humans to yield active, inactive, or toxic metabolites. Although the capacity of the microbiome to modulate drug metabolism has long been known, recent studies have explored these interactions on a much broader scale and have revealed an unprecedented scope of microbial drug metabolism. The implication of this work is that we might be able to predict the capacity of an individual's microbiome to metabolize drugs and use this information to avoid toxicity and inform proper dosing. Here, we provide a tutorial of how to study the microbiome in the context of drug metabolism, focusing on in vitro, rodent, and human studies. We then highlight some limitations and opportunities for the field.
© 2022 The Authors. Clinical and Translational Science published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
The authors declared no competing interests for this work.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

