Integrative biology of persister cell formation: molecular circuitry, phenotypic diversification and fitness effects
- PMID: 36099930
- PMCID: PMC9470271
- DOI: 10.1098/rsif.2022.0129
Integrative biology of persister cell formation: molecular circuitry, phenotypic diversification and fitness effects
Abstract
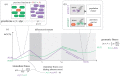
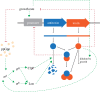
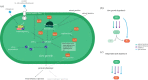
Microbial populations often contain persister cells, which reduce the extinction risk upon sudden stresses. Persister cell formation is deeply intertwined with physiology. Due to this complexity, it cannot be satisfactorily understood by focusing only on mechanistic, physiological or evolutionary aspects. In this review, we take an integrative biology perspective to identify common principles of persister cell formation, which might be applicable across evolutionary-distinct microbes. Persister cells probably evolved to cope with a fundamental trade-off between cellular stress and growth tasks, as any biosynthetic resource investment in growth-supporting proteins is at the expense of stress tasks and vice versa. Natural selection probably favours persister cell subpopulation formation over a single-phenotype strategy, where each cell is prepared for growth and stress to a suboptimal extent, since persister cells can withstand harsher environments and their coexistence with growing cells leads to a higher fitness. The formation of coexisting phenotypes requires bistable molecular circuitry. Bistability probably emerges from growth-modulated, positive feedback loops in the cell's growth versus stress control network, involving interactions between sigma factors, guanosine pentaphosphate and toxin-antitoxin (TA) systems. We conclude that persister cell formation is most likely a response to a sudden reduction in growth rate, which can be achieved by antibiotic addition, nutrient starvation, sudden stresses, nutrient transitions or activation of a TA system.
Keywords: mathematical models; microbial fitness and bet hedging; persister cell microbiology; systems biology.
Figures

References
-
- Bigger J. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. The Lancet 244, 497-500. (10.1016/S0140-6736(00)74210-3) - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

