Stem cell-derived synthetic embryos self-assemble by exploiting cadherin codes and cortical tension
- PMID: 36100738
- PMCID: PMC9481465
- DOI: 10.1038/s41556-022-00984-y
Stem cell-derived synthetic embryos self-assemble by exploiting cadherin codes and cortical tension
Erratum in
-
Author Correction: Stem cell-derived synthetic embryos self-assemble by exploiting cadherin codes and cortical tension.Nat Cell Biol. 2023 Jun;25(6):917. doi: 10.1038/s41556-023-01157-1. Nat Cell Biol. 2023. PMID: 37221392 Free PMC article. No abstract available.
Abstract
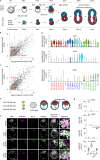
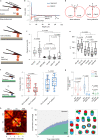
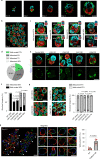
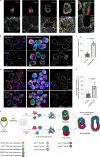
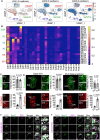
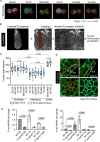
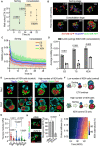
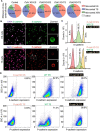
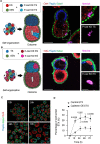
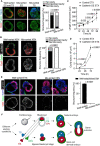
Mammalian embryos sequentially differentiate into trophectoderm and an inner cell mass, the latter of which differentiates into primitive endoderm and epiblast. Trophoblast stem (TS), extraembryonic endoderm (XEN) and embryonic stem (ES) cells derived from these three lineages can self-assemble into synthetic embryos, but the mechanisms remain unknown. Here, we show that a stem cell-specific cadherin code drives synthetic embryogenesis. The XEN cell cadherin code enables XEN cell sorting into a layer below ES cells, recapitulating the sorting of epiblast and primitive endoderm before implantation. The TS cell cadherin code enables TS cell sorting above ES cells, resembling extraembryonic ectoderm clustering above epiblast following implantation. Whereas differential cadherin expression drives initial cell sorting, cortical tension consolidates tissue organization. By optimizing cadherin code expression in different stem cell lines, we tripled the frequency of correctly formed synthetic embryos. Thus, by exploiting cadherin codes from different stages of development, lineage-specific stem cells bypass the preimplantation structure to directly assemble a postimplantation embryo.
© 2022. The Author(s).
Conflict of interest statement
We would like to disclose that we have filed a patent for this study. The applicants and inventors for this Patent are Min Bao and Magdalena Zernicka-Goetz. The patent was filed on September 2, 2022 by Caltech. This patent pertains to and covers the “Differential adhesion and tension guided formation of stem cell derived embryos”. The Patent was filed under the following number: 63/403685. The remaining authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

