Involvement of cell shape and lipid metabolism in glioblastoma resistance to temozolomide
- PMID: 36100765
- PMCID: PMC9958008
- DOI: 10.1038/s41401-022-00984-6
Involvement of cell shape and lipid metabolism in glioblastoma resistance to temozolomide
Abstract
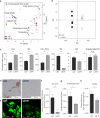
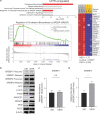
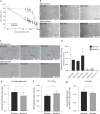
Temozolomide (TMZ) has been used as standard-of-care for glioblastoma multiforme (GBM), but the resistance to TMZ develops quickly and frequently. Thus, more studies are needed to elucidate the resistance mechanisms. In the current study, we investigated the relationship among the three important phenotypes, namely TMZ-resistance, cell shape and lipid metabolism, in GBM cells. We first observed the distinct difference in cell shapes between TMZ-sensitive (U87) and resistant (U87R) GBM cells. We then conducted NMR-based lipid metabolomics, which revealed a significant increase in cholesterol and fatty acid synthesis as well as lower lipid unsaturation in U87R cells. Consistent with the lipid changes, U87R cells exhibited significantly lower membrane fluidity. The transcriptomic analysis demonstrated that lipid synthesis pathways through SREBP were upregulated in U87R cells, which was confirmed at the protein level. Fatostatin, an SREBP inhibitor, and other lipid pathway inhibitors (C75, TOFA) exhibited similar or more potent inhibition on U87R cells compared to sensitive U87 cells. The lower lipid unsaturation ratio, membrane fluidity and higher fatostatin sensitivity were all recapitulated in patient-derived TMZ-resistant primary cells. The observed ternary relationship among cell shape, lipid composition, and TMZ-resistance may be applicable to other drug-resistance cases. SREBP and fatostatin are suggested as a promising target-therapeutic agent pair for drug-resistant glioblastoma.
Keywords: SREBP; cell shape; fatostatin; glioblastoma; lipid metabolism; temozolomide resistance.
© 2022. The Author(s), under exclusive licence to Shanghai Institute of Materia Medica, Chinese Academy of Sciences and Chinese Pharmacological Society.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

