What Is behind the Correlation Analysis of Diarrheagenic E. coli Pathotypes?
- PMID: 36101385
- PMCID: PMC9311887
- DOI: 10.3390/biology11071004
What Is behind the Correlation Analysis of Diarrheagenic E. coli Pathotypes?
Abstract
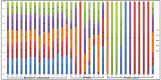
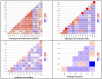
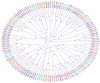
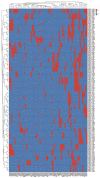
The treatment failure recorded among patients and animals infected with diarrheagenic Escherichia coli (DEC) was increased due to the presence of specific virulence markers among these strains. These markers were used to classify DEC into several pathotypes. We analyzed the correlations between DEC pathotypes and antimicrobial resistances, the existence of virulence genes, serotypes, and hosts. The ETEC pathotype was detected with a high prevalence rate (25%). Moreover, the ETEC and EPEC pathotypes were highly associated with human infections in contrast to the EIEC and EAEC phenotypes, which were commonly recognized among animal isolates. Interestingly, the antimicrobial resistance was affected by E. coli pathotypes. With the exception of EIEC and STEC, imipenem represented the most effective antibiotic against the other pathotypes. There were fixed correlations between the DEC pathotypes and the presence of virulence markers and hosts; meanwhile, their correlation with serotypes was variable. Additionally, the vast majority of our isolates were highly diverse, based on both phenotypic and ERIC molecular typing techniques. Our promising results gave a clear indication for the heterogeneity and weak clonality of DEC pathotypes in Egypt, which can be utilized in the evaluation of the current therapeutic protocols and infection control guidelines.
Keywords: E. coli; clonality; correlation; hosts; pathotypes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Elfaky M.A., Abdel-Hamid M.I., Khalifa E., Alshareef W.A., Mosbah R.A., Elazab S.T., Ghoneim M.M., Al-Sanea M.M., Bendary M.M. Innovative next-generation therapies in combating multi-drug-resistant and multi-virulent Escherichia coli isolates: Insights from in vitro, in vivo, and molecular docking studies. Appl. Microbiol. Biotechnol. 2022;106:1691–1703. doi: 10.1007/s00253-022-11781-w. - DOI - PubMed
-
- Mosallam F.M., Helmy E.A., Bendary M.M., El-Batal I.A. Potency of a novel synthesized Ag- eugenol nanoemulsion for treating some bacterial and fungal pathogens. J. Mater. Res. 2021;36:1524–1537. doi: 10.1557/s43578-021-00226-1. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

