Approaches in Gene Coexpression Analysis in Eukaryotes
- PMID: 36101400
- PMCID: PMC9312353
- DOI: 10.3390/biology11071019
Approaches in Gene Coexpression Analysis in Eukaryotes
Abstract
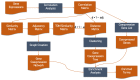
Gene coexpression analysis constitutes a widely used practice for gene partner identification and gene function prediction, consisting of many intricate procedures. The analysis begins with the collection of primary transcriptomic data and their preprocessing, continues with the calculation of the similarity between genes based on their expression values in the selected sample dataset and results in the construction and visualisation of a gene coexpression network (GCN) and its evaluation using biological term enrichment analysis. As gene coexpression analysis has been studied extensively, we present most parts of the methodology in a clear manner and the reasoning behind the selection of some of the techniques. In this review, we offer a comprehensive and comprehensible account of the steps required for performing a complete gene coexpression analysis in eukaryotic organisms. We comment on the use of RNA-Seq vs. microarrays, as well as the best practices for GCN construction. Furthermore, we recount the most popular webtools and standalone applications performing gene coexpression analysis, with details on their methods, features and outputs.
Keywords: RNA-Seq; gene coexpression networks; microarrays; systems biology; transcriptomics; webtool.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- Schneider M.V., Orchard S. Omics Technologies, Data and Bioinformatics Principles. In: Mayer B., editor. Bioinformatics for Omics Data: Methods and Protocols. Humana Press; Totowa, NJ, USA: 2011. pp. 3–30. - PubMed
-
- Usadel B., Obayashi T., Mutwil M., Giorgi F.M., Bassel G.W., Tanimoto M., Chow A., Steinhauser D., Persson S., Provart N.J. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009;32:1633–1651. doi: 10.1111/j.1365-3040.2009.02040.x. - DOI - PubMed
-
- Emamjomeh A., Saboori Robat E., Zahiri J., Solouki M., Khosravi P. Gene co-expression network reconstruction: A review on computational methods for inferring functional information from plant-based expression data. Plant Biotechnol. Rep. 2017;11:71–86. doi: 10.1007/s11816-017-0433-z. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

