A circadian rhythm-related gene signature for predicting relapse risk and immunotherapeutic effect in prostate adenocarcinoma
- PMID: 36103249
- PMCID: PMC9512510
- DOI: 10.18632/aging.204288
A circadian rhythm-related gene signature for predicting relapse risk and immunotherapeutic effect in prostate adenocarcinoma
Abstract
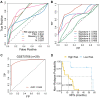
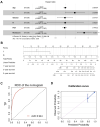
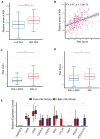
Prostate adenocarcinoma (PRAD) represents the most common male carcinoma in developed countries, its high relapse risk contributes to the second-leading cause of cancer-related deaths. Therefore, it is required to develop an effective signature for predicting the relapse risk of PRAD. To identify a circadian rhythm- (CR-) related predictive signature, we analyzed RNA-seq data of patients with prostate adenocarcinoma (PRAD) from the TCGA and GEO cohort. Seven circadian rhythm- (CR-) related genes (FBXL22, MTA1, TP53, RORC, DRD4, PPARGC1A, ZFHX3) were eventually identified to develop a CR-related signature. AUCs for 3-year overall survival were 0.852, 0.856 and 0.944 in the training set, validation set and an external independent test set (GSE70768), respectively. Kaplan-Meier curve analysis showed that the high-risk group has a reduced relapse-free survival (RFS) than the low-risk group in the training set, validation set, and test set, respectively (P < 0.05). We constructed a prognostic nomogram combining the CR-related signature with T staging to precisely estimate relapse risk of PRAD patients. Finally, we observed that the CR-related gene signature was associated with tumor mutation burden, multiple immune checkpoint molecules and microsatellite instability, and thus could predict response to immune checkpoint inhibitors in PRAD. Conclusively, we developed a circadian rhythm-related gene signature for predicting RFS and immunotherapy efficacy in prostate adenocarcinoma.
Keywords: FBXL22; circadian rhythm; immune checkpoint inhibitor; prognosis; prostate adenocarcinoma.
Conflict of interest statement
Figures






Similar articles
-
A N7-Methylguanine-Related Gene Signature Applicable for the Prognosis and Microenvironment of Prostate Cancer.J Oncol. 2022 May 13;2022:8604216. doi: 10.1155/2022/8604216. eCollection 2022. J Oncol. 2022. PMID: 35602299 Free PMC article.
-
The novel transcriptomic signature of angiogenesis predicts clinical outcome, tumor microenvironment and treatment response for prostate adenocarcinoma.Mol Med. 2022 Jul 14;28(1):78. doi: 10.1186/s10020-022-00504-6. Mol Med. 2022. PMID: 35836112 Free PMC article.
-
Long non-coding RNA profile study identifies an immune-related lncRNA prognostic signature for prostate adenocarcinoma.Int Immunopharmacol. 2021 Dec;101(Pt A):108267. doi: 10.1016/j.intimp.2021.108267. Epub 2021 Nov 2. Int Immunopharmacol. 2021. PMID: 34740081
-
Acircadian rhythm-related gene signature for predicting survival and drug response in HNSC.Front Immunol. 2022 Nov 24;13:1029676. doi: 10.3389/fimmu.2022.1029676. eCollection 2022. Front Immunol. 2022. PMID: 36505439 Free PMC article.
-
Development and validation of novel inflammatory response-related gene signature to predict prostate cancer recurrence and response to immune checkpoint therapy.Math Biosci Eng. 2022 Aug 9;19(11):11345-11366. doi: 10.3934/mbe.2022528. Math Biosci Eng. 2022. PMID: 36124593
Cited by
-
CRS: a circadian rhythm score model for predicting prognosis and treatment response in cancer patients.J Transl Med. 2023 Mar 9;21(1):185. doi: 10.1186/s12967-023-04013-w. J Transl Med. 2023. PMID: 36895015 Free PMC article.
-
DRD4 Interacts with TGF-β Receptors to Drive Colorectal Cancer Metastasis Independently of Dopamine Signaling Pathway.Adv Sci (Weinh). 2025 Feb;12(6):e2413953. doi: 10.1002/advs.202413953. Epub 2024 Dec 16. Adv Sci (Weinh). 2025. PMID: 39679842 Free PMC article.
-
Profiling of a novel circadian clock-related prognostic signature and its role in immune function and response to molecular targeted therapy in pancreatic cancer.Aging (Albany NY). 2023 Jan 9;15(1):119-133. doi: 10.18632/aging.204462. Epub 2023 Jan 9. Aging (Albany NY). 2023. PMID: 36626244 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

