Poly(ADP-ribose) promotes toxicity of C9ORF72 arginine-rich dipeptide repeat proteins
- PMID: 36103513
- PMCID: PMC10359073
- DOI: 10.1126/scitranslmed.abq3215
Poly(ADP-ribose) promotes toxicity of C9ORF72 arginine-rich dipeptide repeat proteins
Abstract
Arginine-rich dipeptide repeat proteins (R-DPRs), abnormal translational products of a GGGGCC hexanucleotide repeat expansion in C9ORF72, play a critical role in C9ORF72-related amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), the most common genetic form of the disorders (c9ALS/FTD). R-DPRs form liquid condensates in vitro, induce stress granule formation in cultured cells, aggregate, and sometimes coaggregate with TDP-43 in postmortem tissue from patients with c9ALS/FTD. However, how these processes are regulated is unclear. Here, we show that loss of poly(ADP-ribose) (PAR) suppresses neurodegeneration in c9ALS/FTD fly models and neurons differentiated from patient-derived induced pluripotent stem cells. Mechanistically, PAR induces R-DPR condensation and promotes R-DPR-induced stress granule formation and TDP-43 aggregation. Moreover, PAR associates with insoluble R-DPR and TDP-43 in postmortem tissue from patients. These findings identified PAR as a promoter of R-DPR toxicity and thus a potential target for treating c9ALS/FTD.
Conflict of interest statement
Figures
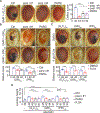

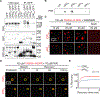
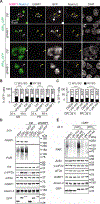


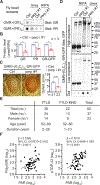
Similar articles
-
Chimeric Peptide Species Contribute to Divergent Dipeptide Repeat Pathology in c9ALS/FTD and SCA36.Neuron. 2020 Jul 22;107(2):292-305.e6. doi: 10.1016/j.neuron.2020.04.011. Epub 2020 May 5. Neuron. 2020. PMID: 32375063 Free PMC article.
-
c-Jun N-Terminal Kinase Promotes Stress Granule Assembly and Neurodegeneration in C9orf72-Mediated ALS and FTD.J Neurosci. 2023 Apr 26;43(17):3186-3197. doi: 10.1523/JNEUROSCI.1799-22.2023. Epub 2023 Apr 4. J Neurosci. 2023. PMID: 37015810 Free PMC article.
-
Arginine-rich dipeptide-repeat proteins as phase disruptors in C9-ALS/FTD.Emerg Top Life Sci. 2020 Dec 11;4(3):293-305. doi: 10.1042/ETLS20190167. Emerg Top Life Sci. 2020. PMID: 32639008 Free PMC article. Review.
-
C9orf72-linked arginine-rich dipeptide repeats aggravate pathological phase separation of G3BP1.Proc Natl Acad Sci U S A. 2024 Dec 10;121(50):e2402847121. doi: 10.1073/pnas.2402847121. Epub 2024 Dec 2. Proc Natl Acad Sci U S A. 2024. PMID: 39621905 Free PMC article.
-
C9orf72 ALS-FTD: recent evidence for dysregulation of the autophagy-lysosome pathway at multiple levels.Autophagy. 2021 Nov;17(11):3306-3322. doi: 10.1080/15548627.2021.1872189. Epub 2021 Feb 26. Autophagy. 2021. PMID: 33632058 Free PMC article. Review.
Cited by
-
PARPs and ADP-ribosylation-mediated biomolecular condensates: determinants, dynamics, and disease implications.Trends Biochem Sci. 2025 Mar;50(3):224-241. doi: 10.1016/j.tibs.2024.12.013. Epub 2025 Feb 7. Trends Biochem Sci. 2025. PMID: 39922741 Review.
-
Poly(GR) interacts with key stress granule factors promoting its assembly into cytoplasmic inclusions.Cell Rep. 2023 Aug 29;42(8):112822. doi: 10.1016/j.celrep.2023.112822. Epub 2023 Jul 19. Cell Rep. 2023. PMID: 37471224 Free PMC article.
-
How villains are made: The translation of dipeptide repeat proteins in C9ORF72-ALS/FTD.Gene. 2023 Mar 30;858:147167. doi: 10.1016/j.gene.2023.147167. Epub 2023 Jan 6. Gene. 2023. PMID: 36621656 Free PMC article. Review.
-
Parthanatos: Mechanisms, modulation, and therapeutic prospects in neurodegenerative disease and stroke.Biochem Pharmacol. 2024 Oct;228:116174. doi: 10.1016/j.bcp.2024.116174. Epub 2024 Mar 27. Biochem Pharmacol. 2024. PMID: 38552851 Review.
-
Transient Poly(ADP-Ribose) Triggers FUS Condensation Hysteresis via a Prion-Like Mechanism.bioRxiv [Preprint]. 2025 Jul 5:2025.07.03.659157. doi: 10.1101/2025.07.03.659157. bioRxiv. 2025. PMID: 40631075 Free PMC article. Preprint.
References
-
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson M, Finch NCA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P,Hsiung G-YR, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). - PMC - PubMed
-
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten J, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M; Consortium ITALSGEN, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy, Singleton J, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor J, A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). - PMC - PubMed
-
- Mori K, Arzberger T, Grässer FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M, van Broeckhoven C, Kremmer E, Kretzschmar HA, Haass C, Edbauer D, Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 126, 881–893 (2013). - PubMed
-
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, van Broeckhoven C, Haass C, Edbauer D, The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013). - PubMed
-
- Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook E, Subramony SH, Ostrow LW, Rothstein JD, Troncoso JC, Ranum LPW, RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U.S.A. 110, E4968–E4977 (2013). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS085207/NS/NINDS NIH HHS/United States
- R01 GM104135/GM/NIGMS NIH HHS/United States
- RF1 NS121143/NS/NINDS NIH HHS/United States
- R01 NS099320/NS/NINDS NIH HHS/United States
- P40 OD018537/OD/NIH HHS/United States
- K99 NS123242/NS/NINDS NIH HHS/United States
- P01 NS084974/NS/NINDS NIH HHS/United States
- TALBOT-MUTIHAC/APR15/832-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- P30 NS050274/NS/NINDS NIH HHS/United States
- R01 NS094239/NS/NINDS NIH HHS/United States
- R37 NS067525/NS/NINDS NIH HHS/United States
- R01 NS082563/NS/NINDS NIH HHS/United States
- U24 NS078736/NS/NINDS NIH HHS/United States
- P01 NS099114/NS/NINDS NIH HHS/United States
- R01 NS117461/NS/NINDS NIH HHS/United States
- R01 NS121125/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

