Eprenetapopt triggers ferroptosis, inhibits NFS1 cysteine desulfurase, and synergizes with serine and glycine dietary restriction
- PMID: 36103522
- PMCID: PMC9473576
- DOI: 10.1126/sciadv.abm9427
Eprenetapopt triggers ferroptosis, inhibits NFS1 cysteine desulfurase, and synergizes with serine and glycine dietary restriction
Abstract
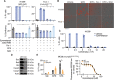
The mechanism of action of eprenetapopt (APR-246, PRIMA-1MET) as an anticancer agent remains unresolved, although the clinical development of eprenetapopt focuses on its reported mechanism of action as a mutant-p53 reactivator. Using unbiased approaches, this study demonstrates that eprenetapopt depletes cellular antioxidant glutathione levels by increasing its turnover, triggering a nonapoptotic, iron-dependent form of cell death known as ferroptosis. Deficiency in genes responsible for supplying cancer cells with the substrates for de novo glutathione synthesis (SLC7A11, SHMT2, and MTHFD1L), as well as the enzymes required to synthesize glutathione (GCLC and GCLM), augments the activity of eprenetapopt. Eprenetapopt also inhibits iron-sulfur cluster biogenesis by limiting the cysteine desulfurase activity of NFS1, which potentiates ferroptosis and may restrict cellular proliferation. The combination of eprenetapopt with dietary serine and glycine restriction synergizes to inhibit esophageal xenograft tumor growth. These findings reframe the canonical view of eprenetapopt from a mutant-p53 reactivator to a ferroptosis inducer.
Figures



References
-
- Bykov V. J. N., Eriksson S. E., Bianchi J., Wiman K. G., Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 18, 89–102 (2018). - PubMed
-
- Cluzeau T., Sebert M., Rahmé R., Cuzzubbo S., Lehmann-Che J., Madelaine I., Peterlin P., Bève B., Attalah H., Chermat F., Miekoutima E., Rauzy O. B., Recher C., Stamatoullas A., Willems L., Raffoux E., Berthon C., Quesnel B., Loschi M., Carpentier A. F., Sallman D. A., Komrokji R., Walter-Petrich A., Chevret S., Ades L., Fenaux P., Eprenetapopt plus azacitidine in TP53-mutated myelodysplastic syndromes and acute myeloid leukemia: A phase II study by the Groupe Francophone des Myelodysplasies (GFM). J. Clin. Oncol. 39, 1575–1583 (2021). - PMC - PubMed
-
- Sallman D. A., DeZern A. E., Garcia-Manero G., Steensma D. P., Roboz G. J., Sekeres M. A., Cluzeau T., Sweet K. L., McLemore A., McGraw K. L., Puskas J., Zhang L., Yao J., Mo Q., Nardelli L., al Ali N. H., Padron E., Korbel G., Attar E. C., Kantarjian H. M., Lancet J. E., Fenaux P., List A. F., Komrokji R. S., Eprenetapopt (APR-246) and azacitidine in TP53-mutant myelodysplastic syndromes. J. Clin. Oncol. 39, 1584–1594 (2021). - PMC - PubMed
-
- Lambert J. M., Gorzov P., Veprintsev D. B., Söderqvist M., Segerbäck D., Bergman J., Fersht A. R., Hainaut P., Wiman K. G., Bykov V. J. N., PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 15, 376–388 (2009). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

