Complex genetics cause and constrain fungal persistence in different parts of the mammalian body
- PMID: 36103708
- PMCID: PMC9630980
- DOI: 10.1093/genetics/iyac138
Complex genetics cause and constrain fungal persistence in different parts of the mammalian body
Abstract
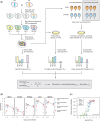
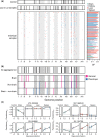
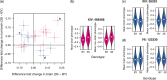
Determining how genetic polymorphisms enable certain fungi to persist in mammalian hosts can improve understanding of opportunistic fungal pathogenesis, a source of substantial human morbidity and mortality. We examined the genetic basis of fungal persistence in mice using a cross between a clinical isolate and the lab reference strain of the budding yeast Saccharomyces cerevisiae. Employing chromosomally encoded DNA barcodes, we tracked the relative abundances of 822 genotyped, haploid segregants in multiple organs over time and performed linkage mapping of their persistence in hosts. Detected loci showed a mix of general and antagonistically pleiotropic effects across organs. General loci showed similar effects across all organs, while antagonistically pleiotropic loci showed contrasting effects in the brain vs the kidneys, liver, and spleen. Persistence in an organ required both generally beneficial alleles and organ-appropriate pleiotropic alleles. This genetic architecture resulted in many segregants persisting in the brain or in nonbrain organs, but few segregants persisting in all organs. These results show complex combinations of genetic polymorphisms collectively cause and constrain fungal persistence in different parts of the mammalian body.
Keywords: antagonistic pleiotropy; complex traits; fungal pathogens; host–pathogen; mammal–fungus interactions; mice; mouse–fungus interactions; yeast.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures


Similar articles
-
Oxidative stress survival in a clinical Saccharomyces cerevisiae isolate is influenced by a major quantitative trait nucleotide.Genetics. 2011 Jul;188(3):709-22. doi: 10.1534/genetics.111.128256. Epub 2011 Apr 21. Genetics. 2011. PMID: 21515583 Free PMC article.
-
Gene-Environment Interactions in Stress Response Contribute Additively to a Genotype-Environment Interaction.PLoS Genet. 2016 Jul 20;12(7):e1006158. doi: 10.1371/journal.pgen.1006158. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27437938 Free PMC article.
-
Comparative polygenic analysis of maximal ethanol accumulation capacity and tolerance to high ethanol levels of cell proliferation in yeast.PLoS Genet. 2013 Jun;9(6):e1003548. doi: 10.1371/journal.pgen.1003548. Epub 2013 Jun 6. PLoS Genet. 2013. PMID: 23754966 Free PMC article.
-
Hidden killers: persistence of opportunistic fungal pathogens in the human host.Curr Opin Microbiol. 2009 Aug;12(4):358-64. doi: 10.1016/j.mib.2009.05.008. Epub 2009 Jun 21. Curr Opin Microbiol. 2009. PMID: 19541532 Review.
-
More evidence for widespread antagonistic pleiotropy in polymorphic disease alleles.Front Genet. 2024 Jun 17;15:1404516. doi: 10.3389/fgene.2024.1404516. eCollection 2024. Front Genet. 2024. PMID: 38952711 Free PMC article. Review.
Cited by
-
Genome-scale analysis of interactions between genetic perturbations and natural variation.Nat Commun. 2024 May 18;15(1):4234. doi: 10.1038/s41467-024-48626-1. Nat Commun. 2024. PMID: 38762544 Free PMC article.
-
Genome-scale analysis of interactions between genetic perturbations and natural variation.bioRxiv [Preprint]. 2024 Jan 16:2023.05.06.539663. doi: 10.1101/2023.05.06.539663. bioRxiv. 2024. Update in: Nat Commun. 2024 May 18;15(1):4234. doi: 10.1038/s41467-024-48626-1. PMID: 38293072 Free PMC article. Updated. Preprint.
-
Global epistasis in budding yeast driven by many natural variants whose effects scale with fitness.bioRxiv [Preprint]. 2025 Jun 1:2025.05.28.656710. doi: 10.1101/2025.05.28.656710. bioRxiv. 2025. Update in: Genetics. 2025 Jul 18:iyaf136. doi: 10.1093/genetics/iyaf136. PMID: 40501638 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

