Epigenetic activation of the FLT3 gene by ZNF384 fusion confers a therapeutic susceptibility in acute lymphoblastic leukemia
- PMID: 36104354
- PMCID: PMC9474531
- DOI: 10.1038/s41467-022-33143-w
Epigenetic activation of the FLT3 gene by ZNF384 fusion confers a therapeutic susceptibility in acute lymphoblastic leukemia
Abstract
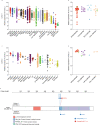
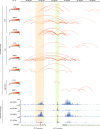
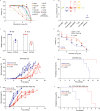
FLT3 is an attractive therapeutic target in acute lymphoblastic leukemia (ALL) but the mechanism for its activation in this cancer is incompletely understood. Profiling global gene expression in large ALL cohorts, we identify over-expression of FLT3 in ZNF384-rearranged ALL, consistently across cases harboring different fusion partners with ZNF384. Mechanistically, we discover an intergenic enhancer element at the FLT3 locus that is exclusively activated in ZNF384-rearranged ALL, with the enhancer-promoter looping directly mediated by the fusion protein. There is also a global enrichment of active enhancers within ZNF384 binding sites across the genome in ZNF384-rearranged ALL cells. Downregulation of ZNF384 blunts FLT3 activation and decreases ALL cell sensitivity to FLT3 inhibitor gilteritinib in vitro. In patient-derived xenograft models of ZNF384-rearranged ALL, gilteritinib exhibits significant anti-leukemia efficacy as a monotherapy in vivo. Collectively, our results provide insights into FLT3 regulation in ALL and point to potential genomics-guided targeted therapy for this patient population.
© 2022. The Author(s).
Conflict of interest statement
J.J.Y. receives a research grant from Takeda Pharmaceutical Company. Other authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

