CRISPR FISHer enables high-sensitivity imaging of nonrepetitive DNA in living cells through phase separation-mediated signal amplification
- PMID: 36104507
- PMCID: PMC9652286
- DOI: 10.1038/s41422-022-00712-z
CRISPR FISHer enables high-sensitivity imaging of nonrepetitive DNA in living cells through phase separation-mediated signal amplification
Erratum in
-
Correction: CRISPR FISHer enables high-sensitivity imaging of nonrepetitive DNA in living cells through phase separation-mediated signal amplification.Cell Res. 2022 Dec;32(12):1133. doi: 10.1038/s41422-022-00742-7. Cell Res. 2022. PMID: 36376418 Free PMC article. No abstract available.
Abstract
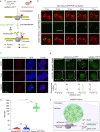
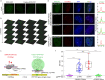
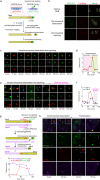
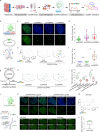
The dynamic three-dimensional structures of chromatin and extrachromosomal DNA molecules regulate fundamental cellular processes and beyond. However, the visualization of specific DNA sequences in live cells, especially nonrepetitive sequences accounting for most of the genome, is still vastly challenging. Here, we introduce a robust CRISPR-mediated fluorescence in situ hybridization amplifier (CRISPR FISHer) system, which exploits engineered sgRNA and protein trimerization domain-mediated, phase separation-based exponential assembly of fluorescent proteins in the CRISPR-targeting locus, conferring enhancements in both local brightness and signal-to-background ratio and thus achieving single sgRNA-directed visualization of native nonrepetitive DNA loci in live cells. In one application, by labeling and tracking the broken ends of chromosomal fragments, CRISPR FISHer enables real-time visualization of the entire process of chromosome breakage, separation, and subsequent intra- or inter-chromosomal ends rejoining in a single live cell. Furthermore, CRISPR FISHer allows the movement of small extrachromosomal circular DNAs (eccDNAs) and invading DNAs to be recorded, revealing substantial differences in dynamic behaviors between chromosomal and extrachromosomal loci. With the potential to track any specified self or non-self DNA sequences, CRISPR FISHer dramatically broadens the scope of live-cell imaging in biological events and for biomedical diagnoses.
© 2022. The author(s).
Conflict of interest statement
C.-Q.S., E.-Z.S., X.-Y.L., and Y.D. are co-inventors of a patent filed by Westlake Laboratory of Life Sciences.
Figures


Similar articles
-
CRISPR-mediated multiplexed live cell imaging of nonrepetitive genomic loci with one guide RNA per locus.Nat Commun. 2022 Apr 6;13(1):1871. doi: 10.1038/s41467-022-29343-z. Nat Commun. 2022. PMID: 35387989 Free PMC article.
-
Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system.Nucleic Acids Res. 2016 May 19;44(9):e86. doi: 10.1093/nar/gkw066. Epub 2016 Feb 4. Nucleic Acids Res. 2016. PMID: 26850639 Free PMC article.
-
CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells.Proc Natl Acad Sci U S A. 2015 Sep 22;112(38):11870-5. doi: 10.1073/pnas.1515692112. Epub 2015 Aug 31. Proc Natl Acad Sci U S A. 2015. PMID: 26324940 Free PMC article.
-
Progress and Challenges for Live-cell Imaging of Genomic Loci Using CRISPR-based Platforms.Genomics Proteomics Bioinformatics. 2019 Apr;17(2):119-128. doi: 10.1016/j.gpb.2018.10.001. Epub 2019 Jan 30. Genomics Proteomics Bioinformatics. 2019. PMID: 30710789 Free PMC article. Review.
-
Imaging Specific Genomic DNA in Living Cells.Annu Rev Biophys. 2016 Jul 5;45:1-23. doi: 10.1146/annurev-biophys-062215-010830. Epub 2016 Apr 27. Annu Rev Biophys. 2016. PMID: 27145877 Free PMC article. Review.
Cited by
-
Sanofi-Cell Research outstanding paper award of 2022.Cell Res. 2023 Dec;33(12):891. doi: 10.1038/s41422-023-00904-1. Cell Res. 2023. PMID: 38017108 Free PMC article. No abstract available.
-
Recruitment and rejoining of remote double-strand DNA breaks for enhanced and precise chromosome editing.Genome Biol. 2025 Mar 11;26(1):53. doi: 10.1186/s13059-025-03523-8. Genome Biol. 2025. PMID: 40069752 Free PMC article.
-
CRISPR-broad: combined design of multi-targeting gRNAs and broad, multiplex target finding.Sci Rep. 2023 Nov 12;13(1):19717. doi: 10.1038/s41598-023-46212-x. Sci Rep. 2023. PMID: 37953351 Free PMC article.
-
HPV integration generates a cellular super-enhancer which functions as ecDNA to regulate genome-wide transcription.Nucleic Acids Res. 2023 May 22;51(9):4237-4251. doi: 10.1093/nar/gkad105. Nucleic Acids Res. 2023. PMID: 36864748 Free PMC article.
-
Live genome imaging by CRISPR engineering: progress and problems.Exp Mol Med. 2025 Jul;57(7):1392-1399. doi: 10.1038/s12276-025-01498-x. Epub 2025 Jul 31. Exp Mol Med. 2025. PMID: 40745002 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

