Glucose-driven TOR-FIE-PRC2 signalling controls plant development
- PMID: 36104568
- PMCID: PMC9530021
- DOI: 10.1038/s41586-022-05171-5
Glucose-driven TOR-FIE-PRC2 signalling controls plant development
Abstract
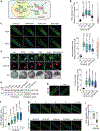
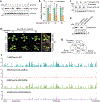
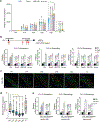
Nutrients and energy have emerged as central modulators of developmental programmes in plants and animals1-3. The evolutionarily conserved target of rapamycin (TOR) kinase is a master integrator of nutrient and energy signalling that controls growth. Despite its key regulatory roles in translation, proliferation, metabolism and autophagy2-5, little is known about how TOR shapes developmental transitions and differentiation. Here we show that glucose-activated TOR kinase controls genome-wide histone H3 trimethylation at K27 (H3K27me3) in Arabidopsis thaliana, which regulates cell fate and development6-10. We identify FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), an indispensable component of Polycomb repressive complex 2 (PRC2), which catalyses H3K27me3 (refs. 6-8,10-12), as a TOR target. Direct phosphorylation by TOR promotes the dynamic translocation of FIE from the cytoplasm to the nucleus. Mutation of the phosphorylation site on FIE abrogates the global H3K27me3 landscape, reprogrammes the transcriptome and disrupts organogenesis in plants. Moreover, glucose-TOR-FIE-PRC2 signalling modulates vernalization-induced floral transition. We propose that this signalling axis serves as a nutritional checkpoint leading to epigenetic silencing of key transcription factor genes that specify stem cell destiny in shoot and root meristems and control leaf, flower and silique patterning, branching and vegetative-to-reproduction transition. Our findings reveal a fundamental mechanism of nutrient signalling in direct epigenome reprogramming, with broad relevance for the developmental control of multicellular organisms.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures


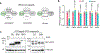







Comment in
-
Plant biology: Identification of the connecTOR linking metabolism, epigenetics and development.Curr Biol. 2022 Nov 21;32(22):R1272-R1274. doi: 10.1016/j.cub.2022.10.002. Curr Biol. 2022. PMID: 36413970
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

