AIM2 upregulation promotes metastatic progression and PD-L1 expression in lung adenocarcinoma
- PMID: 36104978
- PMCID: PMC9807530
- DOI: 10.1111/cas.15584
AIM2 upregulation promotes metastatic progression and PD-L1 expression in lung adenocarcinoma
Abstract
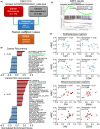
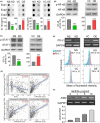
Cancer metastasis leading to the dysfunction of invaded organs is the main cause of the reduced survival rates in lung cancer patients. However, the molecular mechanism for lung cancer metastasis remains unclear. Recently, the increased activity of inflammasome appeared to correlate with the metastatic progression and immunosuppressive ability of various cancer types. Our results showed that the mRNA levels of absence in melanoma 2 (AIM2), one of the inflammasome members, are extensively upregulated in primary tumors compared with normal tissues derived from the TCGA lung adenocarcinoma (LUAD) database. Moreover, Kaplan-Meier analysis demonstrated that a higher mRNA level of AIM2 refers to a poor prognosis in LUAD patients. Particularly, AIM2 upregulation is closely correlated with smoking history and the absence of EGFR/KRAS/ALK mutations in LUAD. We further showed that the endogenous mRNA levels of AIM2 are causally associated with the metastatic potentials of the tested LUAD cell lines. AIM2 knockdown suppressed but overexpression promoted the migration ability and lung colony-forming ability of tested LUAD cells. In addition, we found that AIM2 upregulation is closely associated with an increased level of immune checkpoint gene set, as well as programmed cell death-ligand 1 (PD-L1) transcript, in TCGA LUAD samples. AIM2 knockdown predominantly repressed but overexpression enhanced PD-L1 expression via altering the activity of PD-L1 transcriptional regulators NF-κB/STAT1 in LUAD cells. Our results not only provide a possible mechanism underlying the AIM2-promoted metastatic progression and immune evasion of LUAD but also offer a new strategy for combating metastatic/immunosuppressive LUAD via targeting AIM2 activity.
Keywords: AIM2 inflammasome; PD-L1; cancer metastasis; epithelial-mesenchymal transition; lung cancer.
© 2022 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Figures





References
-
- Araujo A, Ribeiro R, Azevedo I, et al. Genetic polymorphisms of the epidermal growth factor and related receptor in non‐small cell lung cancer – a review of the literature. Oncologist. 2007;12:201‐210. - PubMed
-
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer‐related inflammation. Nature. 2008;454:436‐444. - PubMed
-
- Bremnes RM, Al‐Shibli K, Donnem T, et al. The role of tumor‐infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: emphasis on non‐small cell lung cancer. J Thorac Oncol. 2011;6:824‐833. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

