Mutations responsible for the carbapenemase activity of SME-1
- PMID: 36105999
- PMCID: PMC9377157
- DOI: 10.1039/d2ra02849b
Mutations responsible for the carbapenemase activity of SME-1
Abstract
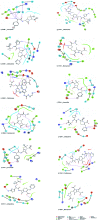
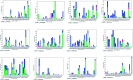
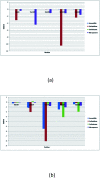
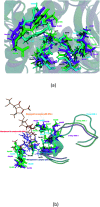
SME-1 is a carbapenemase, produced by Serratia marcescens organism and causes nosocomial infections such as in bloodstream, wounds, urinary tract, or respiratory tract infections. Treatment of such infections becomes very complex due its resistance towards penicillins, cephalosporins, monobactams, and carbapenems. Resistance to such antibiotics is of great medical concern. The misuse and overuse of these antibiotics result in the clinical mutation and production of novel β-lactamase enzymes such as SME-1, which show resistance to carbapenems. Class A contains most of the clinically significant extended spectrum of β-lactamase enzymes and carbapenemases. In this study, class A β-lactamase SME-1 sequence, structure, and binding were compared with naturally mutated class A β-lactamase enzymes and a wild-type TEM-1. This study was performed for revealing mutations, which could be responsible for the carbapenemase activity of SME-1. The dynamic characteristics of SME-1 enzymes manifest a different degree of conservation and variability, which confers them to possess carbapenemase activities. Met69Cys, Glu104Tyr, Tyr105His, Ala237Ser, and Gly238Cys mutations occur in SME-1 as compared to wild-type TEM-1. These mutated residues are present close to active site residues such as Ser70, Lys73, Ser130, Asn132, Glu166, and Asn170, which participate in the hydrolytic reaction of β-lactam antibiotics. Furthermore, these mutated residues demonstrate altered interactions with the β-lactam antibiotics (results in altered binding) and within themselves (results in active site structure alterations), which results in expanding the spectrum of activity of these enzymes. This study provides important insights into the structure and activity relationship of SME-1 enzymes. This is evident from the Ω-loop structure modification, which forms the wall of the active site and repositioning of residues involved in hydrolytic reactions, when present in the complex with meropenem in a stable state of MD simulation at 50 ns. Hence, Met69Cys, Glu104Tyr, Tyr105His, Ala237Ser, and Gly238Cys mutations could result in an altered active site structure, binding, and activity of SME-1 with meropenem and thus become resistantant against meropenem, which is a carbapenem.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures






References
LinkOut - more resources
Full Text Sources
Miscellaneous

