In silico studies of Mpro and PLpro from SARS-CoV-2 and a new class of cephalosporin drugs containing 1,2,4-thiadiazole
- PMID: 36106095
- PMCID: PMC9463509
- DOI: 10.1007/s11224-022-02036-5
In silico studies of Mpro and PLpro from SARS-CoV-2 and a new class of cephalosporin drugs containing 1,2,4-thiadiazole
Abstract
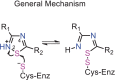
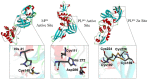
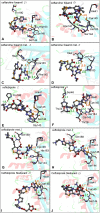
The SARS-CoV-2 proteases Mpro and PLpro are important targets for the development of antivirals against COVID-19. The functional group 1,2,4-thiadiazole has been indicated to inhibit cysteinyl proteases, such as papain and cathepsins. Of note, the 1,2,4-thiadiazole moiety is found in a new class of cephalosporin FDA-approved antibiotics: ceftaroline fosamil, ceftobiprole, and ceftobiprole medocaril. Here we investigated the interaction of these new antibiotics and their main metabolites with the SARS-CoV-2 proteases by molecular docking, molecular dynamics (MD), and density functional theory (DFT) calculations. Our results indicated the PLpro enzyme as a better in silico target for the new antibacterial cephalosporins. The results with ceftaroline fosamil and the dephosphorylate metabolite compounds should be tested as potential inhibitor of PLpro, Mpro, and SARS-CoV-2 replication in vitro. In addition, the data here reported can help in the design of new potential drugs against COVID-19 by exploiting the S atom reactivity in the 1,2,4-thiadiazole moiety.
Supplementary information: The online version contains supplementary material available at 10.1007/s11224-022-02036-5.
Keywords: 1,2,4 thiadiazoles; Cephalosporins; Computational docking; DFT calculations; Drug repurposing; Molecular dynamics.
© The Author(s) 2022.
Conflict of interest statement
Conflict of interestThe authors declare no competing interests.
Figures


Similar articles
-
In Silico and In Vitro Studies of the Approved Antibiotic Ceftaroline Fosamil and Its Metabolites as Inhibitors of SARS-CoV-2 Replication.Viruses. 2025 Mar 28;17(4):491. doi: 10.3390/v17040491. Viruses. 2025. PMID: 40284934 Free PMC article.
-
In silico study to identify novel potential thiadiazole-based molecules as anti-Covid-19 candidates by hierarchical virtual screening and molecular dynamics simulations.Struct Chem. 2022;33(5):1727-1739. doi: 10.1007/s11224-022-01985-1. Epub 2022 Jun 15. Struct Chem. 2022. PMID: 35729938 Free PMC article.
-
Repurposing of FDA-approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS-CoV-2 papain-like protease.J Biomol Struct Dyn. 2021 Sep;39(14):5129-5136. doi: 10.1080/07391102.2020.1784291. Epub 2020 Jun 29. J Biomol Struct Dyn. 2021. PMID: 32597315 Free PMC article.
-
Evaluation of the anti-diabetic drug sitagliptin as a novel attenuate to SARS-CoV-2 evidence-based in silico: molecular docking and molecular dynamics.3 Biotech. 2022 Dec;12(12):344. doi: 10.1007/s13205-022-03406-w. Epub 2022 Nov 7. 3 Biotech. 2022. PMID: 36382241 Free PMC article. Review.
-
In silico molecular docking, dynamics simulation and repurposing of some VEGFR-2 inhibitors based on the SARS-CoV-2-main-protease inhibitor N3.J Biomol Struct Dyn. 2023 Nov;41(19):9267-9281. doi: 10.1080/07391102.2022.2148000. Epub 2022 Nov 18. J Biomol Struct Dyn. 2023. PMID: 36399002 Review.
Cited by
-
In Silico and In Vitro Studies of the Approved Antibiotic Ceftaroline Fosamil and Its Metabolites as Inhibitors of SARS-CoV-2 Replication.Viruses. 2025 Mar 28;17(4):491. doi: 10.3390/v17040491. Viruses. 2025. PMID: 40284934 Free PMC article.
-
The Role of Five-Membered Heterocycles in the Molecular Structure of Antibacterial Drugs Used in Therapy.Pharmaceutics. 2023 Oct 29;15(11):2554. doi: 10.3390/pharmaceutics15112554. Pharmaceutics. 2023. PMID: 38004534 Free PMC article. Review.
References
-
- Berlin DA, Gulick RM, Martinez FJ (2020) Severe Covid-19. 383:2451–2460. 10.1056/NEJMCP2009575 - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
