Reference genome of the Woolly Sculpin, Clinocottus analis
- PMID: 36107748
- PMCID: PMC10019020
- DOI: 10.1093/jhered/esac055
Reference genome of the Woolly Sculpin, Clinocottus analis
Abstract
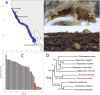
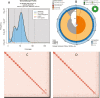
Sculpins (Family Cottidae) are generally cold-temperate intertidal reef fishes most commonly found in the North Pacific. As part of the California Conservation Genomics Project (CCGP), we sequenced the genome of the Woolly Sculpin, Clinocottus analis, to establish a genomic model for understanding phylogeographic structure of inshore marine taxa along the California coast. These patterns, in turn, should further inform the design of marine protected areas using dispersal models based on genomic data. The small genome of C. analis is typical of marine fishes at less than 1 Gb (genome size = 538 Mb), and our assembly is near-chromosome level (contig N50 = 9.1 Mb, scaffold N50 = 21 Mb, BUSCO completeness = 97.9%). Within the context of the CCGP, the Woolly Sculpin genome will be used as a reference for future whole-genome resequencing projects aimed at enhancing our knowledge of the population structure of the species, and efficacy of marine protected areas across the state.
Keywords: CCGP; California Conservation Genomics Project; marine protected areas.
© The Author(s) 2022. Published by Oxford University Press on behalf of The American Genetic Association. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Buser TJ, Andrés López J.. Molecular phylogenetics of sculpins of the subfamily Oligocottinae (Cottidae). Mol Phylogenet Evol. 2015;86:64–74. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

