Global 5'-UTR RNA structure regulates translation of a SERPINA1 mRNA
- PMID: 36107773
- PMCID: PMC9508835
- DOI: 10.1093/nar/gkac739
Global 5'-UTR RNA structure regulates translation of a SERPINA1 mRNA
Abstract
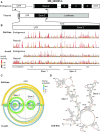
SERPINA1 mRNAs encode the protease inhibitor α-1-antitrypsin and are regulated through post-transcriptional mechanisms. α-1-antitrypsin deficiency leads to chronic obstructive pulmonary disease (COPD) and liver cirrhosis, and specific variants in the 5'-untranslated region (5'-UTR) are associated with COPD. The NM_000295.4 transcript is well expressed and translated in lung and blood and features an extended 5'-UTR that does not contain a competing upstream open reading frame (uORF). We show that the 5'-UTR of NM_000295.4 folds into a well-defined multi-helix structural domain. We systematically destabilized mRNA structure across the NM_000295.4 5'-UTR, and measured changes in (SHAPE quantified) RNA structure and cap-dependent translation relative to a native-sequence reporter. Surprisingly, despite destabilizing local RNA structure, most mutations either had no effect on or decreased translation. Most structure-destabilizing mutations retained native, global 5'-UTR structure. However, those mutations that disrupted the helix that anchors the 5'-UTR domain yielded three groups of non-native structures. Two of these non-native structure groups refolded to create a stable helix near the translation initiation site that decreases translation. Thus, in contrast to the conventional model that RNA structure in 5'-UTRs primarily inhibits translation, complex folding of the NM_000295.4 5'-UTR creates a translation-optimized message by promoting accessibility at the translation initiation site.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
-
- Brantly M., Nukiwa T., Crystal R.G.. Molecular basis of alpha-1-antitrypsin deficiency. Am. J. Med. 1988; 84:13–31. - PubMed
-
- Brantly M. α1-antitrypsin: not just an antiprotease: extending the half-life of a natural anti-inflammatory molecule by conjugation with polyethylene glycol. Am. J. Respir. Cell Mol. Biol. 2002; 27:652–654. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

