Pan-cancer analysis of co-occurring mutations in RAD52 and the BRCA1-BRCA2-PALB2 axis in human cancers
- PMID: 36107942
- PMCID: PMC9477347
- DOI: 10.1371/journal.pone.0273736
Pan-cancer analysis of co-occurring mutations in RAD52 and the BRCA1-BRCA2-PALB2 axis in human cancers
Abstract
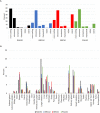
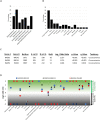
In human cells homologous recombination (HR) is critical for repair of DNA double strand breaks (DSBs) and rescue of stalled or collapsed replication forks. HR is facilitated by RAD51 which is loaded onto DNA by either BRCA2-BRCA1-PALB2 or RAD52. In human culture cells, double-knockdowns of RAD52 and genes in the BRCA1-BRCA2-PALB2 axis are lethal. Mutations in BRCA2, BRCA1 or PALB2 significantly impairs error free HR as RAD51 loading relies on RAD52 which is not as proficient as BRCA2-BRCA1-PALB2. RAD52 also facilitates Single Strand Annealing (SSA) that produces intra-chromosomal deletions. Some RAD52 mutations that affect the SSA function or decrease RAD52 association with DNA can suppress certain BRCA2 associated phenotypes in breast cancers. In this report we did a pan-cancer analysis using data reported on the Catalogue of Somatic Mutations in Cancers (COSMIC) to identify double mutants between RAD52 and BRCA1, BRCA2 or PALB2 that occur in cancer cells. We find that co-occurring mutations are likely in certain cancer tissues but not others. However, all mutations occur in a heterozygous state. Further, using computational and machine learning tools we identified only a handful of pathogenic or driver mutations predicted to significantly affect the function of the proteins. This supports previous findings that co-inactivation of RAD52 with any members of the BRCA2-BRCA1-PALB2 axis is lethal. Molecular modeling also revealed that pathogenic RAD52 mutations co-occurring with mutations in BRCA2-BRCA1-PALB2 axis are either expected to attenuate its SSA function or its interaction with DNA. This study extends previous breast cancer findings to other cancer types and shows that co-occurring mutations likely destabilize HR by similar mechanisms as in breast cancers.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

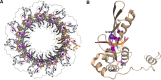

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

