Serial Profiling of Circulating Tumor DNA Identifies Dynamic Evolution of Clinically Actionable Genomic Alterations in High-Risk Neuroblastoma
- PMID: 36108156
- PMCID: PMC9722579
- DOI: 10.1158/2159-8290.CD-22-0287
Serial Profiling of Circulating Tumor DNA Identifies Dynamic Evolution of Clinically Actionable Genomic Alterations in High-Risk Neuroblastoma
Abstract
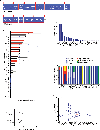
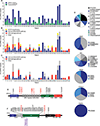
Neuroblastoma evolution, heterogeneity, and resistance remain inadequately defined, suggesting a role for circulating tumor DNA (ctDNA) sequencing. To define the utility of ctDNA profiling in neuroblastoma, 167 blood samples from 48 high-risk patients were evaluated for ctDNA using comprehensive genomic profiling. At least one pathogenic genomic alteration was identified in 56% of samples and 73% of evaluable patients, including clinically actionable ALK and RAS-MAPK pathway variants. Fifteen patients received ALK inhibition (ALKi), and ctDNA data revealed dynamic genomic evolution under ALKi therapeutic pressure. Serial ctDNA profiling detected disease evolution in 15 of 16 patients with a recurrently identified variant-in some cases confirming disease progression prior to standard surveillance methods. Finally, ctDNA-defined ERRFI1 loss-of-function variants were validated in neuroblastoma cellular models, with the mutant proteins exhibiting loss of wild-type ERRFI1's tumor-suppressive functions. Taken together, ctDNA is prevalent in children with high-risk neuroblastoma and should be followed throughout neuroblastoma treatment.
Significance: ctDNA is prevalent in children with neuroblastoma. Serial ctDNA profiling in patients with neuroblastoma improves the detection of potentially clinically actionable and functionally relevant variants in cancer driver genes and delineates dynamic tumor evolution and disease progression beyond that of standard tumor sequencing and clinical surveillance practices. See related commentary by Deubzer et al., p. 2727. This article is highlighted in the In This Issue feature, p. 2711.
©2022 American Association for Cancer Research.
Conflict of interest statement
Conflicts of Interest
M.F., X.L., M.M., M.T., and G.O. are current or past employees of Foundation Medicine. Y.P.M. is a consultant for Pfizer, Inc. No other authors declare any conflict of interest.
Figures





Comment in
-
A Deep Dive into the Circulating ctDNA Cosmos to Vanquish Neuroblastoma.Cancer Discov. 2022 Dec 2;12(12):2727-2729. doi: 10.1158/2159-8290.CD-22-1023. Cancer Discov. 2022. PMID: 36458435
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

