Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75
- PMID: 36108630
- PMCID: PMC9444898
- DOI: 10.1016/j.chom.2022.09.002
Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75
Abstract
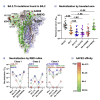
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron subvariant BA.2.75 emerged recently and appears to be spreading. It has nine mutations in spike compared with the currently circulating BA.2, raising concerns that it may further evade vaccine-elicited and therapeutic antibodies. We found BA.2.75 to be moderately more neutralization resistant to sera from vaccinated/boosted individuals than BA.2 (1.8-fold), similar to BA.2.12.1 (1.1-fold), but more neutralization sensitive than BA.4/5 (0.6-fold). Relative to BA.2, BA.2.75 showed heightened resistance to class 1 and class 3 monoclonal antibodies targeting the spike-receptor-binding domain while gaining sensitivity to class 2 antibodies. Resistance was largely conferred by G446S and R460K mutations. BA.2.75 was slightly resistant (3.7-fold) to bebtelovimab, a therapeutic antibody with potent activity against all Omicron subvariants. BA.2.75 also exhibited a higher binding affinity to host receptor ACE2 than other Omicron subvariants. BA.2.75 provides further insight into SARS-CoV-2 evolution as it gains transmissibility while incrementally evading antibody neutralization.
Keywords: ACE2 affinity; BA.2.75; COVID-19; Omicron; SARS-CoV-2; antibody evasion; monoclonal antibodies; serum neutralization; vaccine.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.I., J.Y., Y.H., L.L., and D.D.H. are inventors on patent applications (WO2021236998) or provisional patent applications (63/271,627) filed by Columbia University for a number of SARS-CoV-2 neutralizing antibodies described in this manuscript. Both sets of applications are under review. D.D.H. is a co-founder of TaiMed Biologics and RenBio, consultant to WuXi Biologics and Brii Biosciences, and board director for Vicarious Surgical.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

