Chromosome-scale assemblies of Acanthamoeba castellanii genomes provide insights into Legionella pneumophila infection-related chromatin reorganization
- PMID: 36109147
- PMCID: PMC9528979
- DOI: 10.1101/gr.276375.121
Chromosome-scale assemblies of Acanthamoeba castellanii genomes provide insights into Legionella pneumophila infection-related chromatin reorganization
Abstract
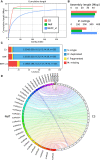
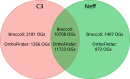
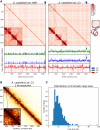
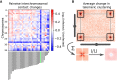
The unicellular amoeba Acanthamoeba castellanii is ubiquitous in aquatic environments, where it preys on bacteria. The organism also hosts bacterial endosymbionts, some of which are parasitic, including human pathogens such as Chlamydia and Legionella spp. Here we report complete, high-quality genome sequences for two extensively studied A. castellanii strains, Neff and C3. Combining long- and short-read data with Hi-C, we generated near chromosome-level assemblies for both strains with 90% of the genome contained in 29 scaffolds for the Neff strain and 31 for the C3 strain. Comparative genomics revealed strain-specific functional enrichment, most notably genes related to signal transduction in the C3 strain and to viral replication in Neff. Furthermore, we characterized the spatial organization of the A. castellanii genome and showed that it is reorganized during infection by Legionella pneumophila Infection-dependent chromatin loops were found to be enriched in genes for signal transduction and phosphorylation processes. In genomic regions where chromatin organization changed during Legionella infection, we found functional enrichment for genes associated with metabolism, organelle assembly, and cytoskeleton organization. Given Legionella infection is known to alter its host's cell cycle, to exploit the host's organelles, and to modulate the host's metabolism in its favor, these changes in chromatin organization may partly be related to mechanisms of host control during Legionella infection.
© 2022 Matthey-Doret et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
